Newsletter
Conditional Knockout Mouse Models
Applied StemCell’s PrimeCKO™ Conditional Knockout (CKO) Mouse Models service allows you to have precise control over where or when your gene of interest in knocked out. Using CRISPR and the Cre-Lox system, we can generate floxed Conditional Knockout Mouse Models for a variety of temporal and spatial knock-out conditions in mice. You can breed the “floxed” PrimeCKO™ conditional knockout mice with commercially available Cre mouse lines, or we can generate a custom Cre mouse model for high-level Cre expression.
- Affordable! Low prices and founder guarantee
- F1 breeding for germline transmission
- Novel CRISPR strategy for higher efficiency and success rate
- Animal IP belongs to customers
- Fast turnaround of 5-8 months
- Free consultation
- Downstream assay services - available
- We also offer a repository of off-shelf mouse models
Our comprehensive service includes vector construction, pronuclear injection, and embryo transfer. The offspring are genotyped for integration, and we deliver at least 2 founders (or germ transmitted F1s). Schedule your free consultation today to discuss your conditional knockout mouse project with one of our experts.
Other Available Mouse Generation Services
- Knockout Mouse Model Generation
- Knockin Mouse Model Generation
- Reporter Mouse Model Generation
- Rose26 Knockin Mouse Model
- Point Mutation Mouse Model Generation
- CRISPR Humanized Mouse Model Generation Service
- Transgenic Mice
Products and Services
Case Studies
Case Study 1
A conditional knock-out mouse model with LoxP sequences inserted in intron 1 and downstream of 3’ UTR of the desired locus.
This conditional knockout mouse model was generated using CRISPR Technology by inserting LoxP sequences in intron 1 and downstream of 3’ UTR of the gene of interest . In the first step, a mixture of active guide RNA molecules (gRNAs), two single stranded oligo donor nucleotide (ssODN) and qualified Cas-9 mRNA was prepared and injected into cytoplasm of C57BL/6 embryos. The second step was to screen new mice born from the microinjection for the presence of LoxP sites at designated locations using PCR. And the third step was to confirm the potentially positive animals by sequencing the modified regions in the mouse genomic locus.
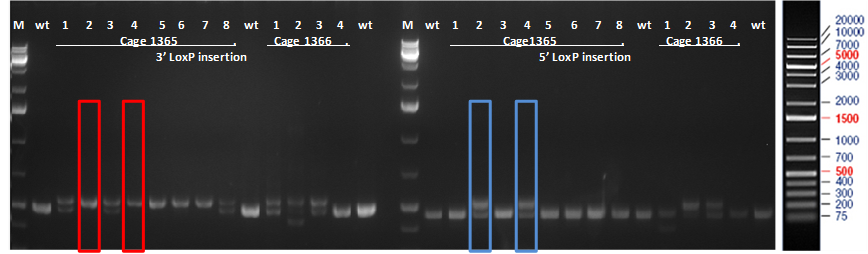
Figure 1. PCR results of mice born after microinjection of the embryos with CRISPR cocktail. Two out of twelve mice were identified as founders and showed the expected fragment shifts for both 5’ and 3’ LoxP insertions. A LoxP insertion at the 5’site, or intron 1 produced a 513bp PCR fragment (blue box; WT: 473bp) and LoxP insertion at the 3’-targeting site produced a 539bp PCR fragment (red box; WT: 499 bp).

Figure 2. Representative illustration sequence analyses of founder mice confirms LoxP insertion at 5’ and 3’ location at the desired genome locus.
Case Study 2
Generation of a conditional mouse model with a floxed exon using CRISPR
This conditional knockout mouse (CKO) model was generated using CRISPR technology by inserting two LoxP cassettes on either side of the exon to be conditionally removed. The exon can then be removed by crossbreeding the floxed mouse with a Cre-recombinase-expressing mouse. We generated the floxed model using three well optimized steps: (1) a mixture of two sets of active guide RNA molecules (gRNAs), two single stranded oligodeoxylnucleotides (ssODNs) and qualified Cas-9 mRNA was injected into the cytoplasm of C57BL/6 embryo; (2) new mice born from microinjection were screened using a scheme combining PCR and restriction enzyme digest; (3) we confirmed the floxed allele positive animals by sequencing the modified region in the mouse locus.
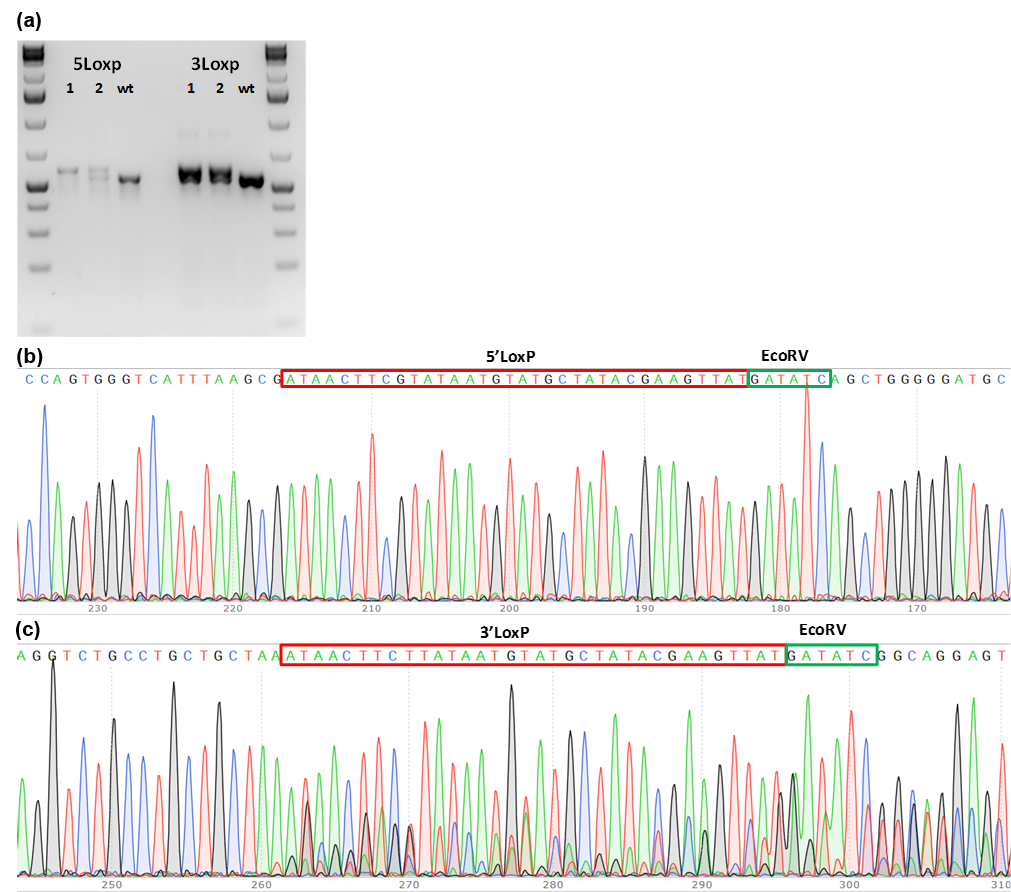
Figure 1. PCR genotype screening of founder mice. (a) PCR product size shift for 5’LoxP and 3’LoxP insertions; (b) Chromatograms of 5’LoxP and 3’LoxP PCR fragments from founder mice. Genomic DNA extracted from individual mice born from microinjection of the embryos were subjected to genotyping PCR. Two mice were identified as potential founders. Sequencing results showed that both mice have LoxP insertions and that mouse# 1 may have 5’ LoxP on both alleles.
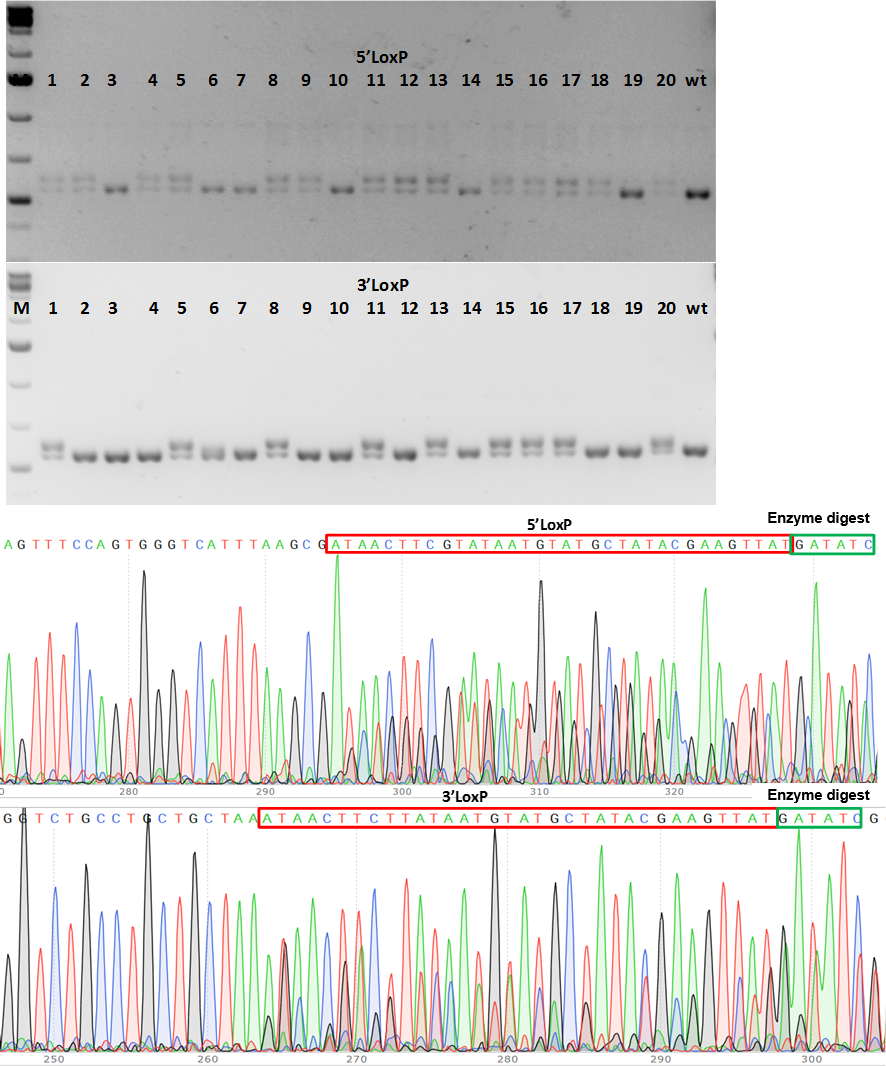
Figure 2. PCR genotype screening of F1 CKO mice. (a) PCR product size shows fragment size shift for 5’LoxP and 3’LoxP insertions in 9 mice (#1, 5, 8, 11, 13, 15; 16, 17 and 20 (b) Chromatograms of 5’LoxP and 3’LoxP sequences at correct location in genome.
Application Notes
CRISPR for Generating Conditional Knockout Mice
The most commonly used conditional knockout (CKO) system is the Cre-LoxP system, where the gene of interest (targeted exons) is flanked by two LoxP sequences (also called floxed allele). The flanking LoxP sequences are inserted at specific sites on either side of the gene of interest using CRISPR technology. The LoxP sites are a target for the Cre Recombinase which catalyzes the deletion of the floxed exon(s).
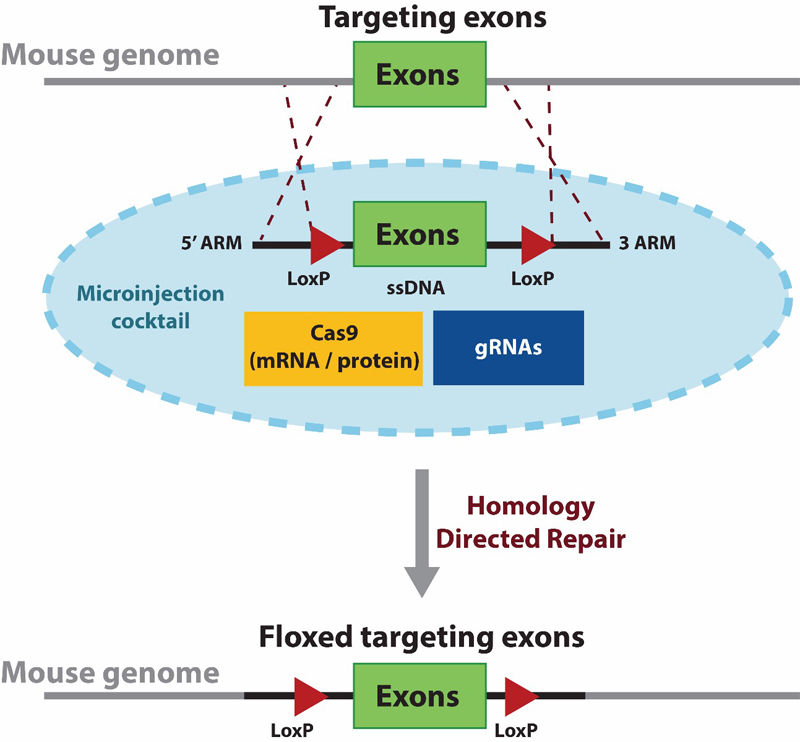
Diagram 1. The schematic describesthe first stage in developing a conditional knock-out mouse model using CRISPR to generate a floxed (loxP flanked exon) mouse. A single stranded donor DNA (ssDNA) is used for delivering the floxed targeting exons to replace the wildtype form. The donor contains two LoxP sequences flanking the targeted exon(s) along with 5' and 3' homologous arms for directing a site-specific homology directed repair. The donor ssDNA is delivered along with Cas9 (mRNA or protein) and validated gRNAs via microinjection.
CKO mice are generated by crossbreeding two transgenic mouse lines, one with homozygous “floxed” (flanked by loxP) allele, and the other bearing Cre recombinase transgene under the control of a promoter directing tissue specific expression or ubiquitous expression. The Cre expression has minimal unwanted effects in the animal as the mouse genome does not contain endogenous loxP sites, providing an ideal background for site-specific recombination.
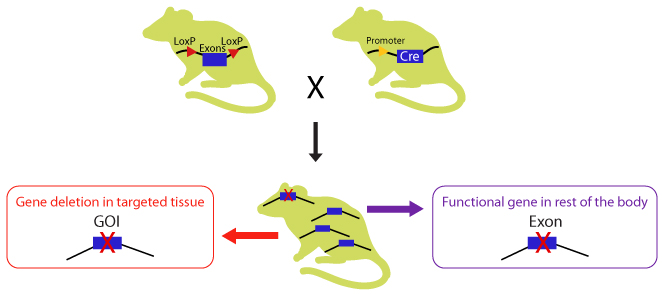
Diagram 2. Crossbreeding the CKO mouse with a Cre-recombinase expressing mouse. The Cre expression is driven by a promoter of choice: a tissue specific or ubiquitous promoter. The expressed Cre recombinase deletes the floxed exon(s) in a tissue specific manner there by causing a frameshift in downstream sequence.
Support Materials
Posters:
Publications
CRISPR Mouse/ Rat Models: Knock-in, Knockout, and Conditional Knockout
CRISPR Technology
-
Smalley, E. (2016). CRISPR mouse model boom, rat model renaissance. Nature Biotechnology. 34, 893–894.
-
Baker, M. (2014). Gene editing at CRISPR speed. Nature biotechnology, 32(4), 309-313.
CRISPR Knock-in H11 Locus in Pigs
-
Ruan, J., Li, H., Xu, K., Wu, T., Wei, J., Zhou, R., ... & Chen-Tsai, R. Y. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Scientific reports, 5, 14253.
Knock-in, Knockout, Conditional Knock-out
- Park, J., Jung, E., Lee, S. H., & Chung, W. S. (2020). CDC50A dependent phosphatidylserine exposure induces inhibitory post-synapse elimination by microglia. bioRxiv.
- Ramachandra Rao, S., Fliesler, S. J., Kotla, P., Nguyen, M. N., & Pittler, S. J. (2020). Lack of Overt Retinal Degeneration in a K42E Dhdds Knock-In Mouse Model of RP59. Cells, 9(4), 896.
- Beurg, M., Barlow, A., Furness, D. N., & Fettiplace, R. (2019). A Tmc1 mutation reduces calcium permeability and expression of mechanoelectrical transduction channels in cochlear hair cells. Proceedings of the National Academy of Sciences, 116(41), 20743-20749.
- Goldring, A. C., Beurg, M., & Fettiplace, R. (2019). The contribution of TMC1 to adaptation of mechanoelectrical transduction channels in cochlear outer hair cells. The Journal of physiology.
- Hwang, S., He, Y., Xiang, X., Seo, W., Kim, S. J., Ma, J., ... & Kunos, G. (2019). Interleukin‐22 ameliorates neutrophil‐driven nonalcoholic steatohepatitis through multiple targets. Hepatology https://doi.org/10.1002/hep.31031.
- Dumesic, P. A., Egan, D. F., Gut, P., Tran, M. T., Parisi, A., Chatterjee, N., ... & Dou, F. (2019). An Evolutionarily Conserved uORF Regulates PGC1α and Oxidative Metabolism in Mice, Flies, and Bluefin Tuna. Cell metabolism.
- Liang, T., Zhang, H., Xu, Q., Wang, S., Qin, C., & Lu, Y. (2019). Mutant Dentin Sialophosphoprotein Causes Dentinogenesis Imperfecta. Journal of dental research, 0022034519854029.
- Qian, W., Miner, C. A., Ingle, H., Platt, D. J., Baldridge, M. T., & Miner, J. J. (2019). A human STAT1 gain-of-function mutation impairs CD8+ T cell responses against gammaherpesvirus-68. Journal of virology, JVI-00307.
- Kweon, S. M., Chen, Y., Moon, E., Kvederaviciutė, K., Klimasauskas, S., & Feldman, D. E. (2019). An Adversarial DNA N6-Methyladenine-Sensor Network Preserves Polycomb Silencing. Molecular Cell. https://doi.org/10.1016/j.molcel.2019.03.018
- Deng, F., He, S., Cui, S., Shi, Y., Tan, Y., Li, Z., ... & Peng, L. (2018). A Molecular Targeted Immunotherapeutic Strategy for Ulcerative Colitis via Dual-Targeting Nanoparticles Delivering miR-146b to Intestinal Macrophages. Journal of Crohn's and Colitis.
- Jo, S., Fonseca, T. L., Bocco, B. M. D. C., Fernandes, G. W., McAninch, E. A., Bolin, A. P., ... & Németh, D. (2018). Type 2 deiodinase polymorphism causes ER stress and hypothyroidism in the brain. The Journal of Clinical Investigation.
- Langston, R. G., Rudenko, I. N., Kumaran, R., Hauser, D. N., Kaganovich, A., Ponce, L. B., ... & Beilina, A. (2018). Differences in Stability, Activity and Mutation Effects Between Human and Mouse Leucine-Rich Repeat Kinase 2. Neurochemical research, 1-14.
- Amara, N., Tholen, M., & Bogyo, M. (2018). Chemical tools for selective activity profiling of endogenously expressed MMP-14 in multicellular models. ACS Chemical Biology. doi: 10.1021/acschembio.8b00562.
- Allocca, S., Ciano, M., Ciardulli, M. C., D’Ambrosio, C., Scaloni, A., Sarnataro, D., ... & Bonatti, S. (2018). An αB-Crystallin Peptide Rescues Compartmentalization and Trafficking Response to Cu Overload of ATP7B-H1069Q, the Most Frequent Cause of Wilson Disease in the Caucasian Population. International journal of molecular sciences, 19(7).
-
*Peng, L., Zhang, H., Hao, Y., Xu, F., Yang, J., Zhang, R., ... & Chen, C. (2016). Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine, 14, 83-96.
-
*Hu, J. K., Crampton, J. C., Locci, M., & Crotty, S. (2016). CRISPR-mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ triple gene disruption reveals NKT cell defects but not T follicular helper cell defects. PloS one, 11(5), e0156074.
-
*Besschetnova, T. Y., Ichimura, T., Katebi, N., Croix, B. S., Bonventre, J. V., & Olsen, B. R. (2015). Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biology, 42, 56-73.
-
*McKenzie, C. W., Craige, B., Kroeger, T. V., Finn, R., Wyatt, T. A., Sisson, J. H., ... & Lee, L. (2015). CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Molecular biology of the cell, 26(18), 3140-3149.
-
*Bishop, K. A., Harrington, A., Kouranova, E., Weinstein, E. J., Rosen, C. J., Cui, X., & Liaw, L. (2016). CRISPR/Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3: Genes, Genomes, Genetics, 6(7), 2051-2061.
For more journal references, please visit our comprehensive list of citations and reference publications.
FAQs
What is the basis of your design algorithm for the sgRNAs?
What is your success rate for generation CRISPR/Cas9 conditional knockout mice?
How would you recommend your customers to screen for off-target effects and has that been done for the conditional mice you have produced?
What is your delivery time and cost for generating a CKO mouse model?
Do you provide guarantee that the KO will succeed?




