Newsletter
iPSC Characterization Services: Check Pluripotency Markers, Chromosomal Abnormality, STR, & WGS
Stringent molecular and functional assays are necessary to evaluate pluripotency and to rule-out genetic aberrations due to reprogramming and stress from in vitro culture in iPSC lines. Loss of genetic integrity can affect desired cellular phenotype and compromise interpretation and translation of results to a clinical setting.
ASC offers comprehensive services needed to completely characterize your human and mouse pluripotent stem cell lines (PSCs):
- Pluripotency and lineage-specific marker immunostaining
- Karyotyping (Chromosome counting, G-banding, array analysis)
- qPCR, RNA-seq
- Trilineage differentiation potential: embryoid body (EB) formation, qRT-PCR
- HLA typing, whole genome sequencing (WGS), single tandem repeat (STR) genotyping, copy number variations (CNV) and more
Products and Services
Technical Details
The new era of induced pluripotent stem cell (iPSCs) technology has vast potential as an in vitro research tool as well as in regenerative medicine. However, there are lot we still need to understand about the iPSCs and what the reprogramming and in vitro differentiation processes. As reprogramming of human iPSCs from somatic cells is becoming increasingly mainstream for both basic and applied research, there is accumulating evidence that rigorous characterization for more than just pluripotency markers and functionality is required. Many times, the reprogramming methods and the stress of culturing the iPSCs in vitro introduces genetic and epigenetic alterations that can change the cellular phenotype and affect the integrity of experiments using these cell lines 1. As well, CRISPR or other genome editing technologies used for engineering cell line models for disease modeling and cell regeneration therapy also tend to introduce undesirable genetic alterations, especially during the single cell cloning stage necessitating the screening of a greater number of clones to generate the desired line 2.
Recently, large biobanks for iPSCs are established to provide a repository (stem cell banks) of standardized, master iPSC lines from different donors: healthy and documented disease mutations, difference ages and ethnicities, and source tissue, in order to support and expand its scientific use and for detailed understanding of its translational applications. It is crucial that these master iPSCs have been fully characterized as per regulatory requirements in order to provide reliable scientific data and preclinical research model for clinical use 3.
In addition to our custom iPSC generation, CRISPR/ Cas9 genome editing and differentiation to CNS and other somatic lineages, Applied StemCell also provides a complete suite of services to characterize your iPSC and ESC line. Some of the routine assays recommended to validate your stem cell lines include:
|
Pluripotency markers |
To characterize iPSCs and ESCs for expression of pluripotency markers with high resolution microscopy and imaging capabilities |
|
|
Karyotyping/ cytogenetics |
To identify and evaluate the stem cell genomic stability, chromosomal abnormalities, and copy number variants (CNVs)
|
|
|
Differentiation potential |
Ability to form three germ layers and differentiate into somatic lineages |
|
|
Microbiological contamination |
To ensure purity of the cells |
|
We also provide advanced characterization of iPSCs and other cell lines to suit your project needs:
|
Cell authentication/ DNA fingerprinting |
To establish purity and identity of cell line
|
|
|
HLA typing |
To identify HLA markers for potential transplantation studies |
|
|
Genome Sequencing |
Using NGS to detect single nucleotide variants |
|
|
Residual programming factors |
To detect presence of reprogramming footprint if using non-integrating methods such as plasmids and retroviruses |
|
It is recommended that some key characterization assays such as karyotype analysis and STR analysis be carried out repeatedly at various passages, before and after genome editing and/or directed differentiation.
References
1. Weissbein, U., Plotnik, O., Vershkov, D., & Benvenisty, N. (2017). Culture-induced recurrent epigenetic aberrations in human pluripotent stem cells. PLoS genetics, 13(8), e1006979.
2. Bai, Q., Ramirez, J. M., Becker, F., Pantesco, V., Lavabre-Bertrand, T., Hovatta, O., ... & De Vos, J. (2014). Temporal analysis of genome alterations induced by single-cell passaging in human embryonic stem cells. Stem cells and development, 24(5), 653-662.
3. Garreta, E., Sanchez, S., Lajara, J., Montserrat, N., & Belmonte, J. C. I. (2018). Roadblocks in the Path of iPSC to the Clinic. Current transplantation reports, 5(1), 14-18.
Case Studies
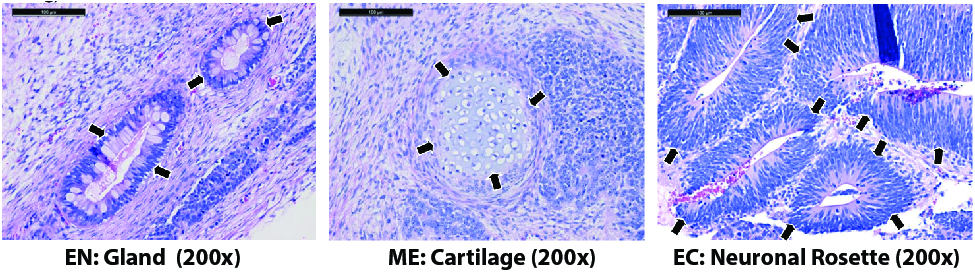
Figure. H&E staining of kidney and testis teratomas from mice injected with the ASE-9203 control iPSC line shows differentiated tissues representing the three germ layers, indicated by arrowheads. EN: endoderm; ME: mesoderm; EC: ectoderm.
Application Notes
Direct Differentiation of Control ‘Master” iPSC Line, ASE-9209 into the Three Germ Layers
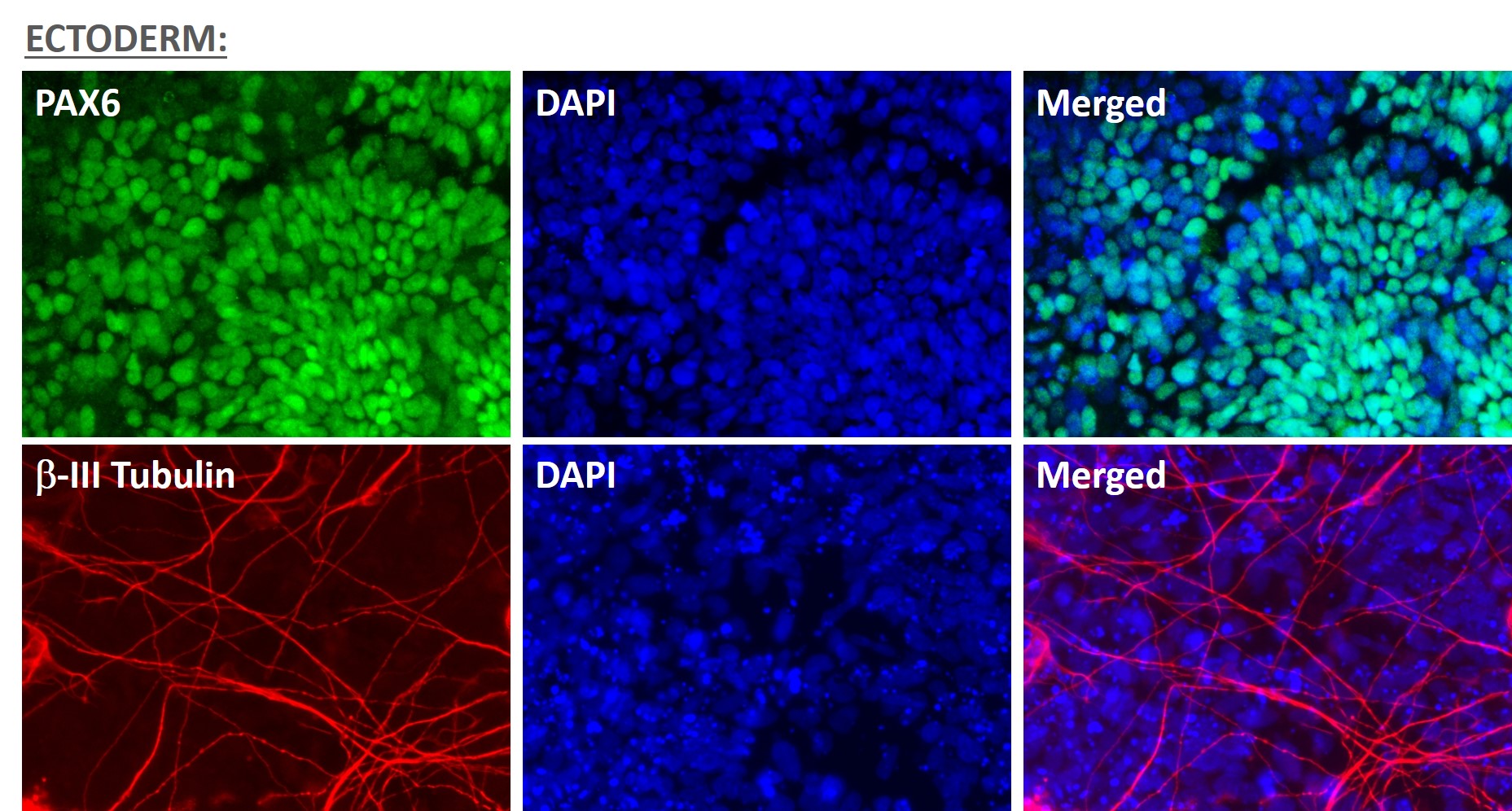
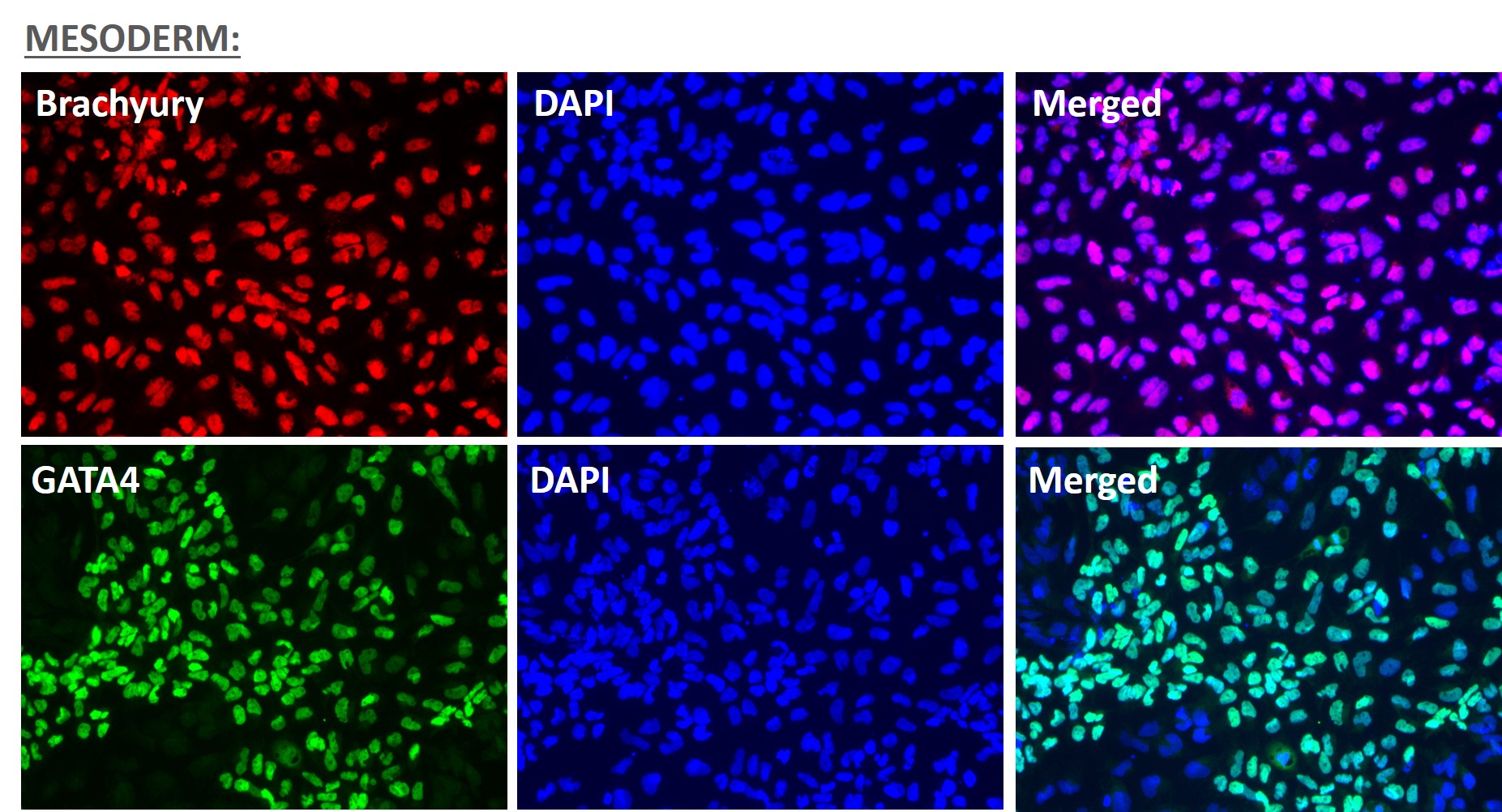
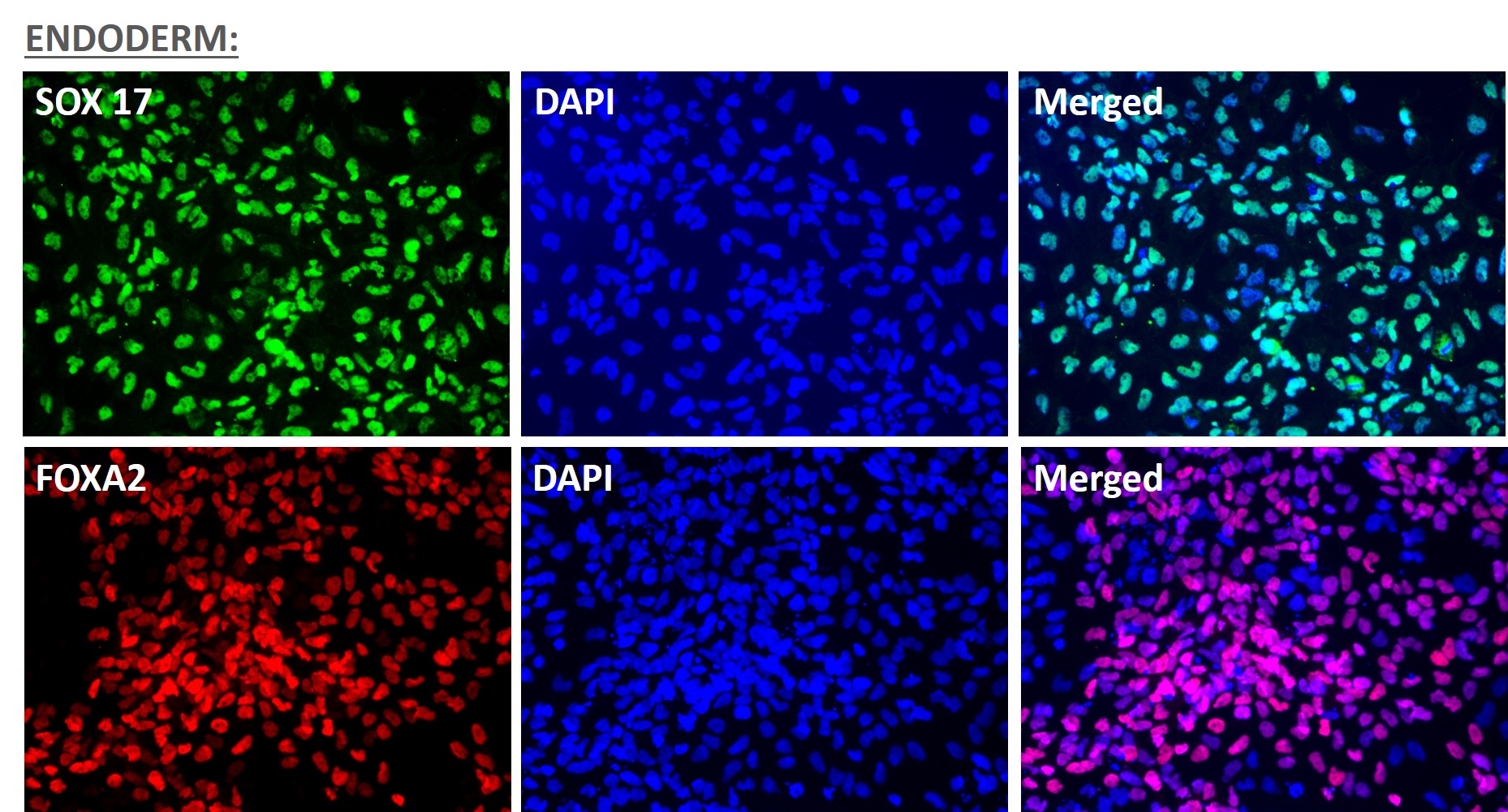
Figure 1. Immunofluorescent staining for lineage-specific biomarkers of three germ layers after direct differentiation of hiPSCs. Control hiPSC line, ASE-9209 (female, fibroblasts) were differentiated to specific lineages of the germ layers using well-established and optimized protocols. Immunostaining for biomarkers of each lineage was performed to confirm lineage commitment. Ectoderm markers: neuronal lineage markers, PAX6 (green), b-III Tubulin (red); Mesoderm markers: Brachyury (green) and GATA4 (red); Endoderm markers: SOX17 (green) and FOXA2 (red); DAPI (blue) was used to stain for nuclear localization.
Support Materials
Brochure:
Publications
Reference Publications (selected cited/published articles)
- Huang, W., Xu, M., Li, R., Baskfield, A., Kouznetsova, J., Beers, J., ... & Zheng, W. (2019). An induced pluripotent stem cell line (TRNDi006-A) from a MPS IIIB patient carrying homozygous mutation of p. Glu153Lys in the NAGLU gene. Stem Cell Research, 101427.
- Li, R., Baskfield, A., Lin, Y., Beers, J., Zou, J., Liu, C., ... & Zheng, W. (2019). Generation of an induced pluripotent stem cell line (TRNDi003-A) from a Noonan syndrome with multiple lentigines (NSML) patient carrying a p. Q510P mutation in the PTPN11 gene. Stem cell research, 34, 101374.
- Li, R., Pradhan, M., Xu, M., Baskfield, A., Farkhondeh, A., Cheng, Y. S., ... & Rodems, S. (2018). Generation of an induced pluripotent stem cell line (TRNDi002-B) from a patient carrying compound heterozygous p. Q208X and p. G310G mutations in the NGLY1 gene. Stem Cell Research, 101362.
- Vozdek, R., Long, Y., & Ma, D. K. (2018). The receptor tyrosine kinase HIR-1 coordinates HIF-independent responses to hypoxia and extracellular matrix injury. Sci. Signal., 11(550), eaat0138.
- Ou, J., Ball, J. M., Luan, Y., Zhao, T., Miyagishima, K. J., Xu, Y., ... & Mallon, B. S. (2018). iPSCs from a Hibernator Provide a Platform for Studying Cold Adaptation and Its Potential Medical Applications. Cell, 173(4), 851-863. https://doi.org/10.1016/j.cell.2018.03.010
- Teves, S. S., An, L., Bhargava-Shah, A., Xie, L., Darzacq, X., & Tjian, R. (2018). A stable mode of bookmarking by TBP recruits RNA Polymerase II to mitotic chromosomes. bioRxiv, 257451. DOI: 10.1101/257451
- Hansen, A. S., Pustova, I., Cattoglio, C., Tjian, R., & Darzacq, X. (2017). CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife, 6.
- Vermilyea, S. C., Guthrie, S., Meyer, M., Smuga-Otto, K., Braun, K., Howden, S., ... & Golos, T. G. (2017). Induced Pluripotent Stem Cell-Derived Dopaminergic Neurons from Adult Common Marmoset Fibroblasts. Stem cells and development, 26(17), 1225-1235. https://doi.org/10.1089/scd.2017.0069.



