Newsletter
Genome Edited iPSC Lines
Applied StemCell offers a research-ready, isogenic panel of genome-edited human iPSCs and their differentiated cells from well-characterized control, parental iPSCs. These gene-edited human iPSCs and differentiated neuronal lineage cell lines are ideal for hard-to-model or hard-to-source neurological diseases disease modeling, neurotoxicity screening, and drug discovery and screening studies.
- Neuronal disease-specific gene knockout lines recapitulate disease phenotype of patient-derived iPSCs
- Safe harbor locus reporter gene knockin iPSC lines available for quantitative screening experiments
- Ready-to-use Differentiated neuronal lineages: astrocytes, neurons, and dopamine neurons
- An isogenic panel of control neurons, dopamine neurons, and astrocytes available as an experimental control
Products and Services
Technical Details
Genome engineering in induced pluripotent stem cells (iPSCs) offers the advantage of generating numerous cell line models in an isogenic background along with the parental cell line as an ideal control, where patient-derived primary cells are difficult to obtain. As well, genome editing to introduce and correct mutations in control and disease iPSC lines, is a powerful tool for comparative study of genetic mechanisms involved in cellular/organ development and disease pathology.
The true potential of the iPSC technology lies in the directed differentiation of iPSCs into a desired cell line lineage, especially into neuronal lineage cells as it provides a more predictive source of central nervous system (CNS) models. These cells are valuable tools for high throughput screening of neurotoxicological and neuroprotective compounds.
Applications:
- Physiologically relevant cell lines to model Parkinson’s disease, Alzheimer’s disease, ALS, Schizophrenia, and Autism.
- Predictive models to study neurogenetic mechanisms
- Suitable for high-throughput drug screening of neuroprotective and neurotoxicological compounds.
We also have disease-specific iPSC lines derived from patients with ALS8, Parkinson’s disease and Diabetes type II patient samples. Please inquire for details on differentiation of these genome-edited cell lines as well as custom differentiation services.
Applied StemCell’s Gene Editing iPSC Lines:
1. Neuronal Disease Specific Isogenic Knockout Lines:
Isogenic Knock-out Lines |
Associated Disease |
Description |
|
PARK2 -/- |
PD |
Biallelic KO |
|
PARK7 -/- |
PD |
Biallelic KO |
|
PINK1 -/- |
PD |
Biallelic KO |
|
LRRK2 -/- |
PD |
Biallelic KO |
|
Park2-/-; Park7-/- |
PD |
Biallelic KO |
|
Park2-/-; Pink1-/- |
PD |
Biallelic KO |
|
APOE -/- |
Alzheimer's disease |
Biallelic KO |
|
SOD1 -/- |
ALS |
Biallelic KO |
|
DISC1 -/- |
Schizophrenia |
Biallelic KO |
|
CNTNAP2 -/- |
Autism |
Biallelic KO |
|
BDNF -/- |
CNS |
Biallelic KO |
2. Lineage-specific Reporter Gene Knock-in Lines:
Knock-in Lineage-specific Reporters |
Description |
|
MAP2-Nanoluc-Halotag KI |
Neuron reporter |
|
GFAP-Nanoluc-Halotag KI |
Astrocyte reporter |
|
MBP-Nanoluc-Halotag KI |
Oligodendrocyte reporter |
3. Safe Harbor Locus Reporter Gene Knock-in Lines:
Safe-harbor knock-in lines |
Description |
|
CAG-GFP, AAVS/Chr19 |
Ubiquitous reporter |
|
DCXp-GFP |
Neuron reporter |
Application Notes
Disease-specific Knockout iPSC Line: APOE Knockout (ASE-9405)
The ASE-9405 iPSC line is engineered with a bi-allelic (homozygous) knockout for APOE gene (APOE-/-) that has been implicated in Alzheimer’s disease etiology. The parental iPSC is ASE-9109 which is an integration-free, normal karyotype iPSC derived from male, CD34+ cord blood cells. The APOE knockout line can be further differentiated into an isogenic panel of neurons and glia for disease modeling and drug/ toxicity screening applications.
1. Genotyping of APOE-/- bi-allelic KO clone F4: APOE KO

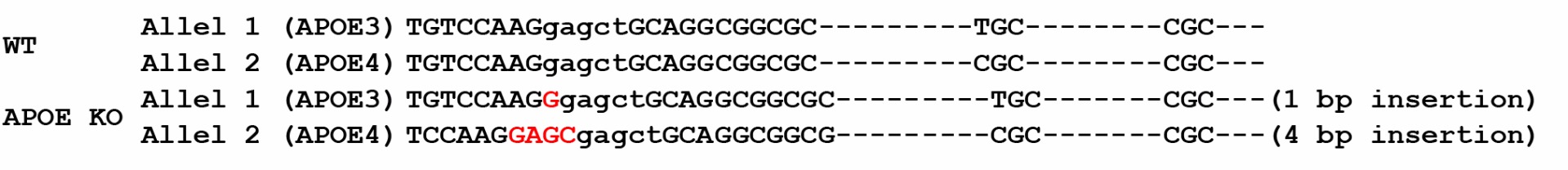
Figure 1. Sequence alignment between parental/ control iPSC line (ASE-9109; male) and APOE-/- clone. The homozygous knockout clone shows a 4 bp deletion in allele 1, a 3 bp deletion in allele 2, and a 4bp insertion in allele 2, as compared to wild type (WT) sequence in parental iPSC line.
2. Expression of Pluripotency Markers
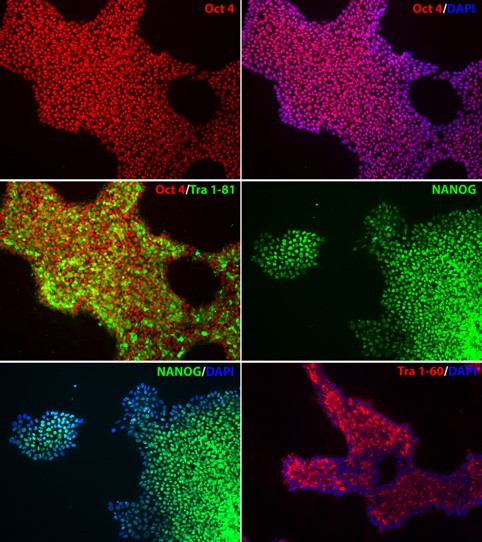
Figure 2. Expression of pluripotency markers in APOE-/- iPSC line. The homozygous knockout iPSC line, APOE-/-, expresses pluripotency markers OCT4, NANOG, TRA-1-60, and TRA-1-81, indicating pluripotency of the iPSC line after genome editing. Nucleus stained with DAPI (blue).
3. Expression of APOE in neural stem cells differentiated from APOE-/- iPSC-line
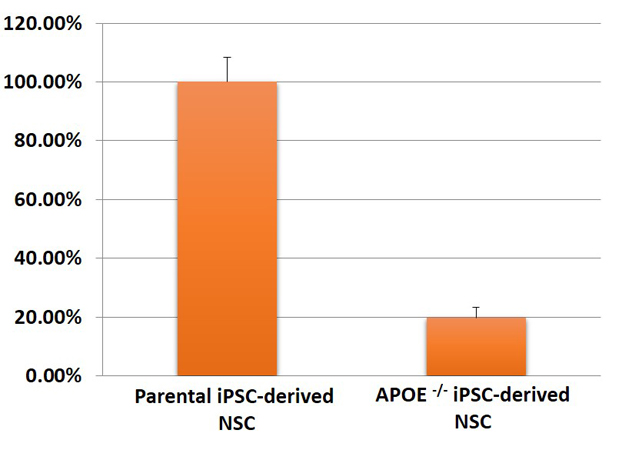
Figure 3. Expression of APOE in neural stem cells (NSC) differentiated from master/ parental iPSC line (ASE-9109) and APOE-/- iPSC line (ASE-9405) was quantified using qPCR. The APOE mRNA was significantly downregulated in the APOE-knockout NSC as compared to expression in the parental/ control iPSC line.
| WT | APOE-/- |
| 1022 | -4 |
Figure 4. APOE Expression in APOE knockout NSCs by microarray. APOE expression in the knockout NSC line was significantly lower than the wild type NSC line.
4. Differentiation of APOE -/- iPSCs into Neurons and GFAP
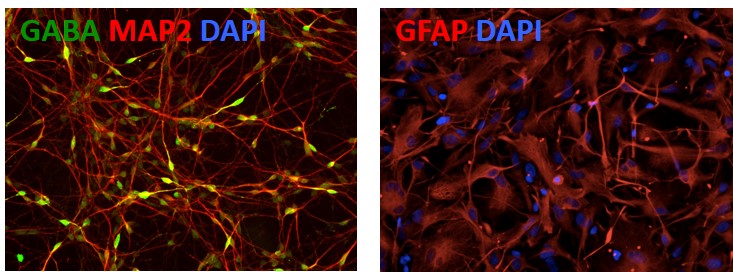
Figure 5. Differentiation of APOE-/- iPSCs into cortical neurons and astrocytes. The APOE-/- line can be differentiated into cortical neurons (GABA; green) and astrocytes (GFAP; red) to model Alzheimer's disease. Other markers: Neuronal marker (MAP2; red) and nucleus staining (DAPI; blue).
iPSC lines with reporter gene inserted at a safe harbor locus: AAVS1-DCXp-GFP iPSC line (ASE-9502)
ASE-9502 iPSC line is a mono-allelic (heterozygous) knock-in iPSC line engineered with a GFP reporter tag driven by a neuronal-lineage-specific promoter, Doublecortin (DCX) inserted into the AAVS safe harbor locus. The parental iPSC is ASE-9109 which is an integration-free, normal karyotype iPSC derived from male, CD34+ cord blood cells. The AAVS-DCX-GFP Knock-in iPSC line can be further differentiated into an isogenic panel of neurons and glia for disease modeling and drug/ toxicity screening applications.
1. Schematic representation of AAVS-DCXp-GFP reporter construct
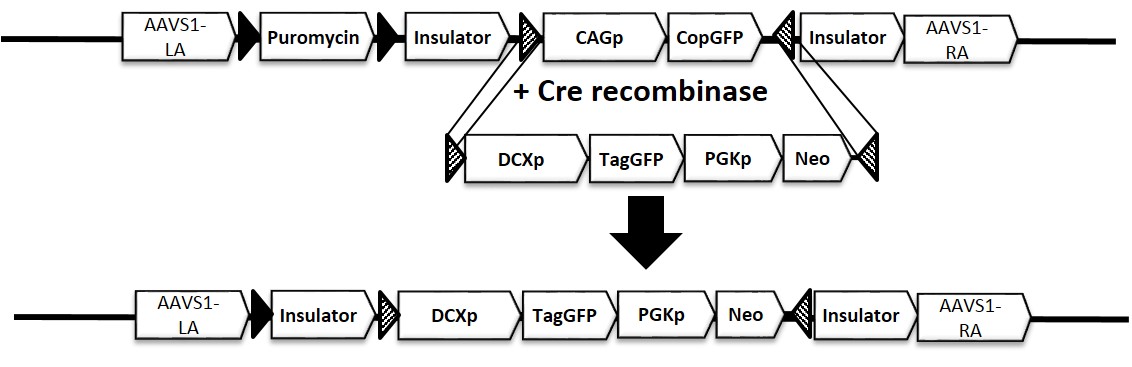
Figure 1. Schematic representation of iPSC line with reporter gene inserted in AAVS safe harbor locus, AAVS1-DCXp-GFP (ASE-9502). The reporter gene expression is driven by a neuronal-lineage (neuronal development) specific promoter, Doublecortin (DCXp). The ASE-9502 cell line is derived from master cell line with a ubiquitous promoter expressing a CopGFP reporter gene. After Cre-recombinase mediated cassette exchange (RMCE), a single copy of the DCXp-TagGFP cassette is inserted into the AAVS1 locus.
2. Validation of neuronal-lineage specific reporter cassette knock-in in AAVS1 safe harbor locus
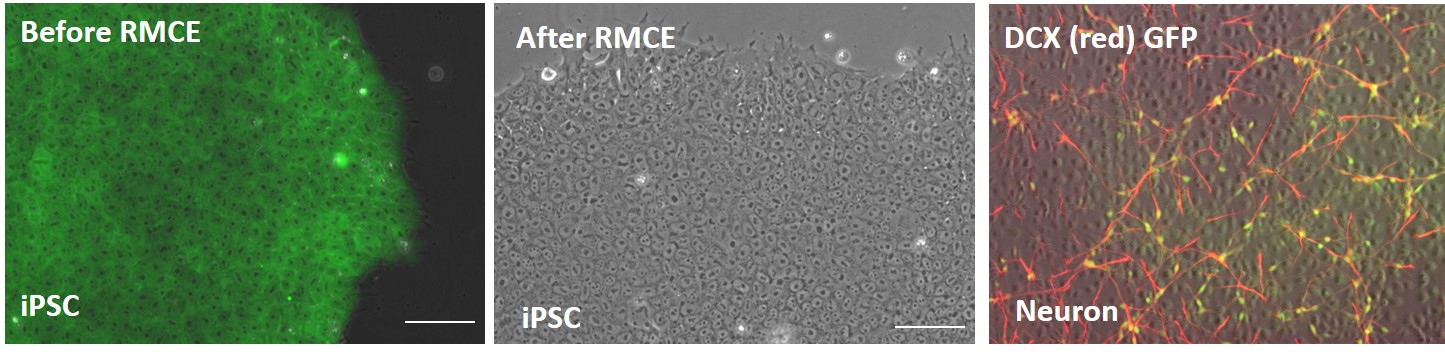
Figure 2. Validation of the DCXp-GFP reporter gene insertion in AAVS1 locus in iPSC line, ASE-9502. In the AAVS-CAG-GFP master cell line, GFP expression is driven by the ubiquitous promoter, CAG (all cells are stained green for GFP). After RMCE, there is no GFP expression in the iPSCs confirming the absence of a ubiquitous CAG promoter after cassette exchange. When these iPSCs are differentiated into neurons, immunohistochemical characterization showed the co-localization (yellow) of GFP (green) and DCXp-positive neurons (red). The GFP is expressed only in neuronal cells confirming the insertion of the reporter gene in the correct locus.
Support Materials
FAQs
How specific is the GFP reporter driven by the DCXp promoter in the AAVS1-DCXp-GFP knock-in iPSC line?
Is the ASE-9045 APOE-/- iPSC line contain knockout of APOE4 or another isoform?




