Newsletter
CRISPR iPSC Gene Knockout
Knockout induced pluripotent stem cells (iPSCs) are powerful and versatile tools that can be used to understand and elucidate phenotype-genotype relationships, the functional role of genes (loss of function), antibody validation, and more in different cell/tissue types. As a premier iPSC gene editing service provider with over 12 years of genetic engineering and stem cell expertise, Applied StemCell (ASC) can efficiently knock out one or more gene(s) of interest (GOI) using our optimized CRISPR/Cas9 protocols.
- 98% project success rate
- Single-cell cloning, precious deletion, select Homozygous or Heterozygous
- High throughput workflows enable up to 60% faster timelines
- We work with your healthy or disease iPSC lines; control iPSC lines available
- Various gene knockout modalities: large knockout, frameshift, fragment excision, stop cassette insertion, double gene knockout, gene fusion with interstitial deletion
- Fully customizable service, add downstream differentiation/assay development or upstream iPSC generation services
- GMP iPSC Gene Editing Available >> Learn More
If you are looking for iPSCs to genetically modify, ASC can generate the iPSCs you need from your human or non-human samples. We can even further differentiate your gene knockout iPSCs to the cell type of your choice, including NK cells, T cells, and more. While we offer fully customizable research-grade iPSC generation, genome editing, and differentiation services, we encourage you to look into our new GMP-grade iPSC service platform to learn more about our new GMP-grade service options!
GMP iPSC Gene Editing & Additional Services Available >> Learn More
CRISPR iPSC Knockout Workflow

Products and Services
Case Studies
Case Study #1: Frameshift Knockout Mutation in a Control Human iPSC Line
Goal: To generate a frameshift knockout (KO) mutation in a control human iPSC line using CRISPR/Cas9 and non-homologous end joining (NHEJ).
Out of two gRNAs designed to target an early exon to generate the desired frameshift mutation, one gRNA was selected based on functional validation for optimal NHEJ frequency. After transfection and clonal selection, individual colonies were screened for desired frameshift mutation by PCR using primer sets for the region flanking the gRNA cut site (Figure 3A) and then confirmed by Sanger sequencing (Figure 3B).
A. 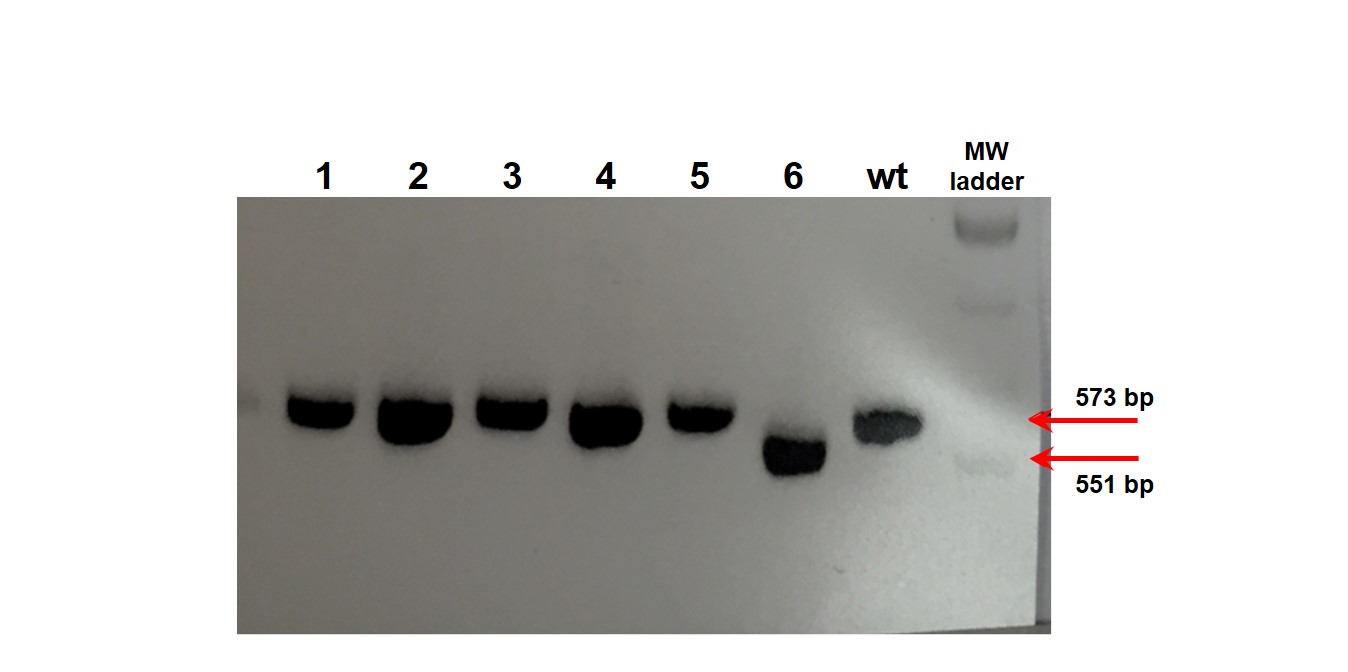
B.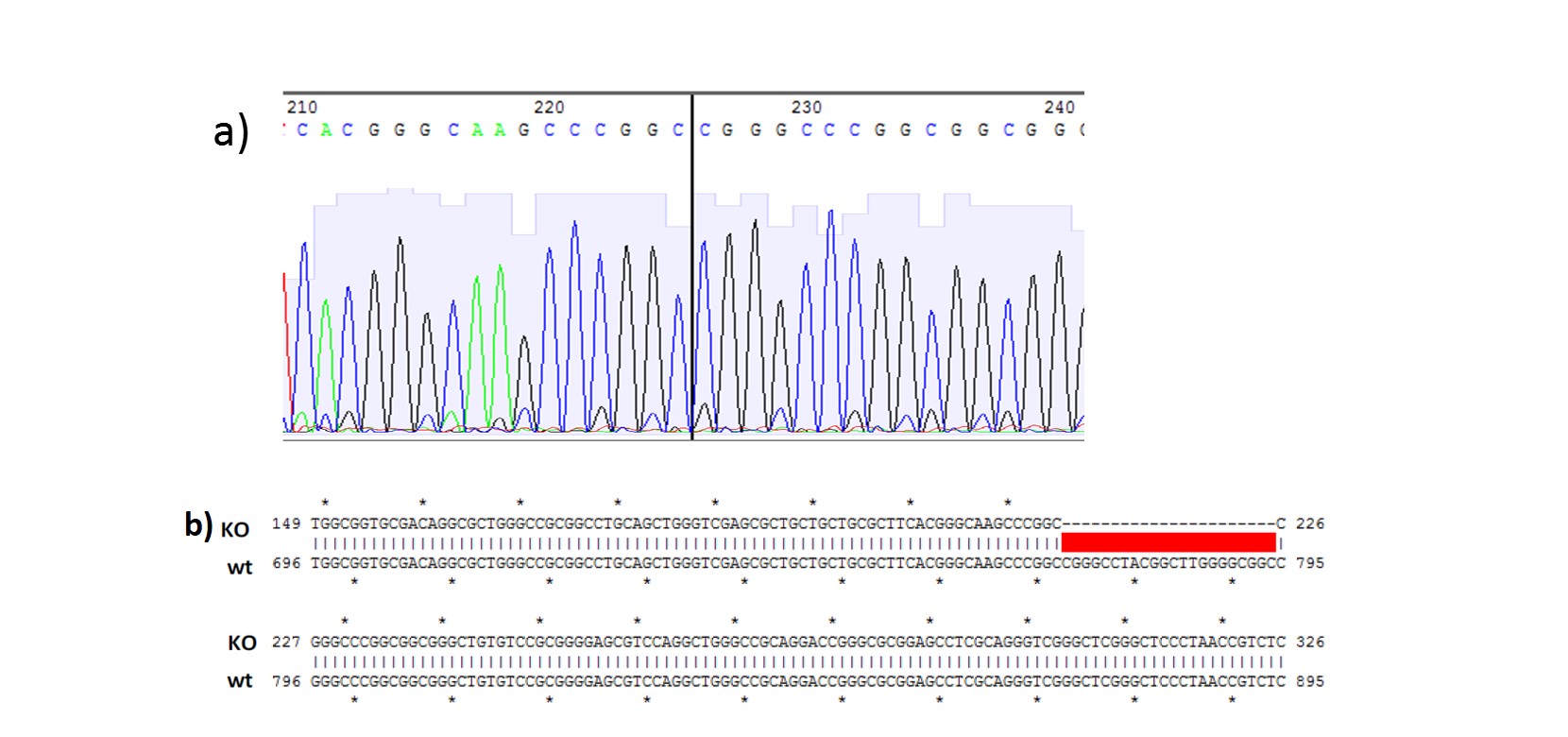
Figure 1. (A) PCR genotyping of KO single cell iPSC clones. PCR amplification products for the region flanking the gRNA cut site were run on a 2% agarose gel. The presence of a 551 bp PCR product identified clone #6 as an iPSC line with a 22bp deletion. Lanes containing the 573 bp PCR product do not have deletion and are similar to wildtype (wt); (B) a) Sequence chromatogram: The vertical line in the histogram indicates the new sequence generated by the 22 bp deletion; b).Sequence alignment between KO clone (#6; top) and wildtype clone (wt; bottom).
Case Study #2: Heterozygous Large Deletion of a Targeted Sequence in Human iPSCs
Goal: To delete a large fragment of DNA (~100kb) from a specific locus in control human iPSCs to mimic a chromosomal abnormality seen in a rare neurodevelopmental disorder.
1. Human iPSCs were obtained from the customer and the cell line was validated for single cell clonability and other parameters.

Figure 1. Schematic representation of the workflow undertaken for the project.
2. Guide RNAs (gRNAs) were designed based on the distance to the modification site and the off-target profile. The gRNAs were cloned into Cas9-expressing plasmids and validated for indel formation in iPSCs. Two gRNAs, upstream and one downstream of targeted regions, were identified after subsequent analysis of the cells by PCR and sequencing to confirm indel formation at the gRNA cutting site.
3. The 2 gRNAs were then co-transfected into iPSCs and single cell clones were isolated. Genomic DNA from the single cell clones was subjected to PCR (figure 2 and 3) using primer pairs for upstream and downstream of the targeted sequence, and verified by Sanger sequencing (not shown). One positive heterozygous clone was identified with the deletion of the ~100 kb sequence at targeted chromosomal locus.
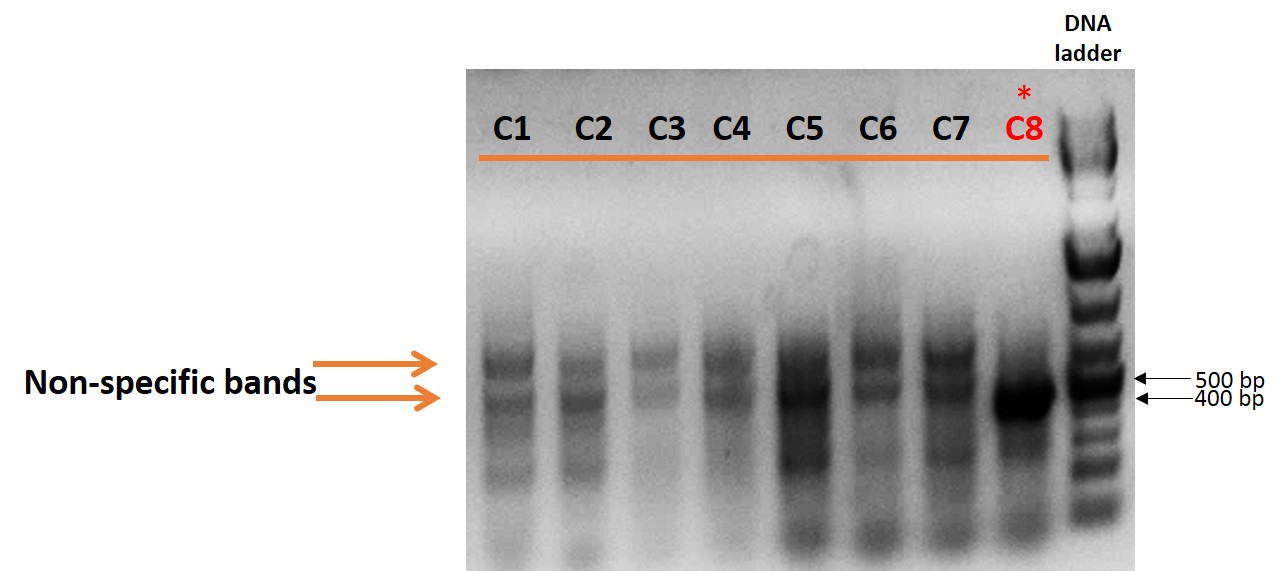
Figure 2. PCR gel electrophoresis to detect knockout of ~100kb fragment in a specific chromosomal locus in human iPSCs, showed a shorter band at the size of 440bp with clone C8. Further sequencing identified this clone (C8) to be positive (*) for the targeted deletion.
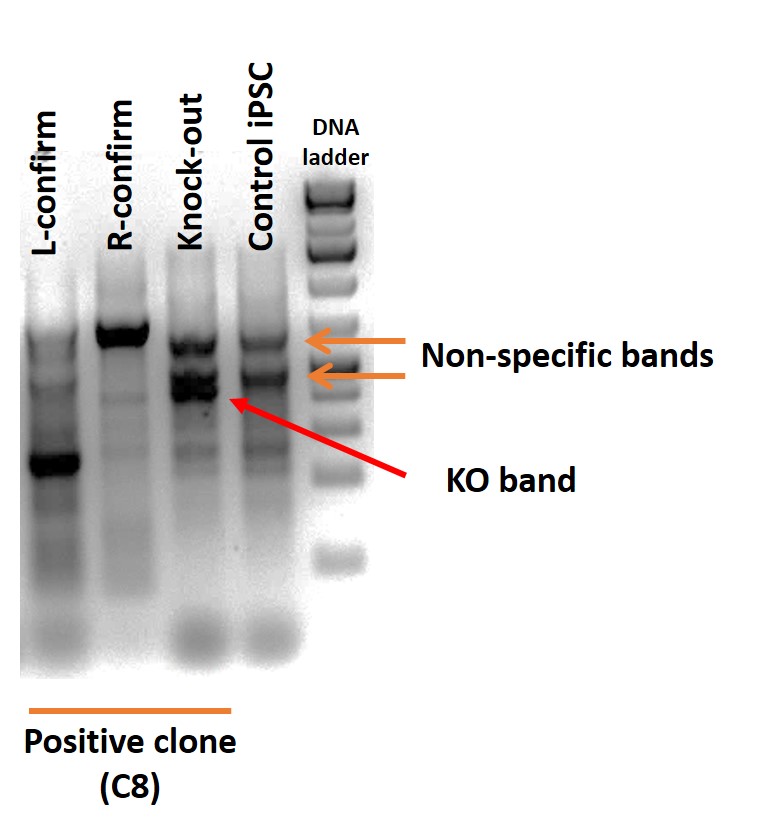
Figure 3. PCR to confirm deletion and to detect homozygous/heterozygous clone with primers flanking the locus of interest: L confirm: upstream forward/reverse primers; R confirm: downstream forward/reverse primers. Clone C8 shows bands for both upstream and downstream PCRs, suggesting that clone C8 is a heterozygous clone.
Support Materials




