Newsletter
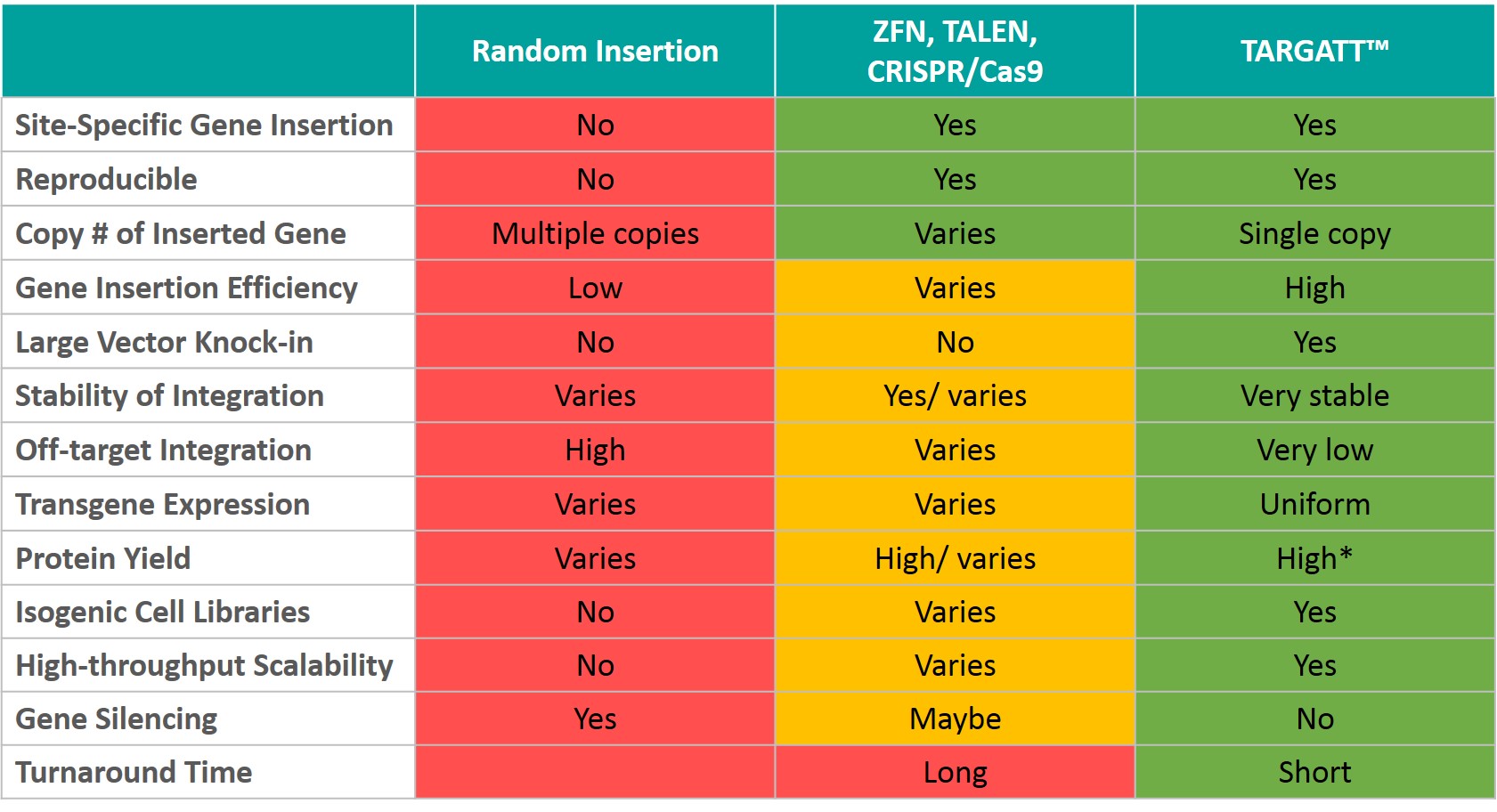
Applied StemCell’s (ASC) proprietary TARGATT™ technology, enables fast and site-specific, stable integration of large DNA fragments (up to 20 kb) into an intergenic, transcriptionally active safe harbor locus with very high efficiency. The preselected locus is engineered to contain an "attP" integrase recognition landing pad where single-copy gene integration occurs when used in conjunction with an “attB” containing donor plasmid and integrase expression.
The TARGATT™ gene editing platform is versatile and can be used for the development of large fragment knock-in cell lines, bioproduction, and library construction. This technology circumvents problems associated with random integration such as position effect, and gene silencing or instability due to the integration of multiple copies of the transgene.
ASC can accurately and efficiently engineer the necessary landing pad into the cell line of your choice. Ready-to-use TARGATT TM Master Cell Lines (iPSC, HEK293, and CHO) are also available for integration of your gene of interest (GOI) at a preselected locus that has been tested for uniformed, high gene expression. Contact us today to schedule your free consultation!




Technology Comparision
We hold exclusive rights from Sandford for the groundbreaking, site-specific knock-in technology, TARGATTTM. TARGATTTM is an efficient, fast system that permits the integration of a large fragment, up to 20 kb, at a pre-selected safe-harbor locus. The single-copy insertion of any gene of interest, including chimeric antigen receptor (CAR) genes, at a transcriptionally active locus, enables the evasion of significant problems that arise from random insertion such as gene interruption. Moreover, it eliminates the position effect and guarantees high-level, uniformed transgene expression.
ASC holds rights to CRISPR/Cas9 technology and is one of the earliest providers of CRISPR services. Throughout the years we have optimized our protocols to deliver high-throughput genome editing services for complex and mainstream genetic engineering of iPSCs.

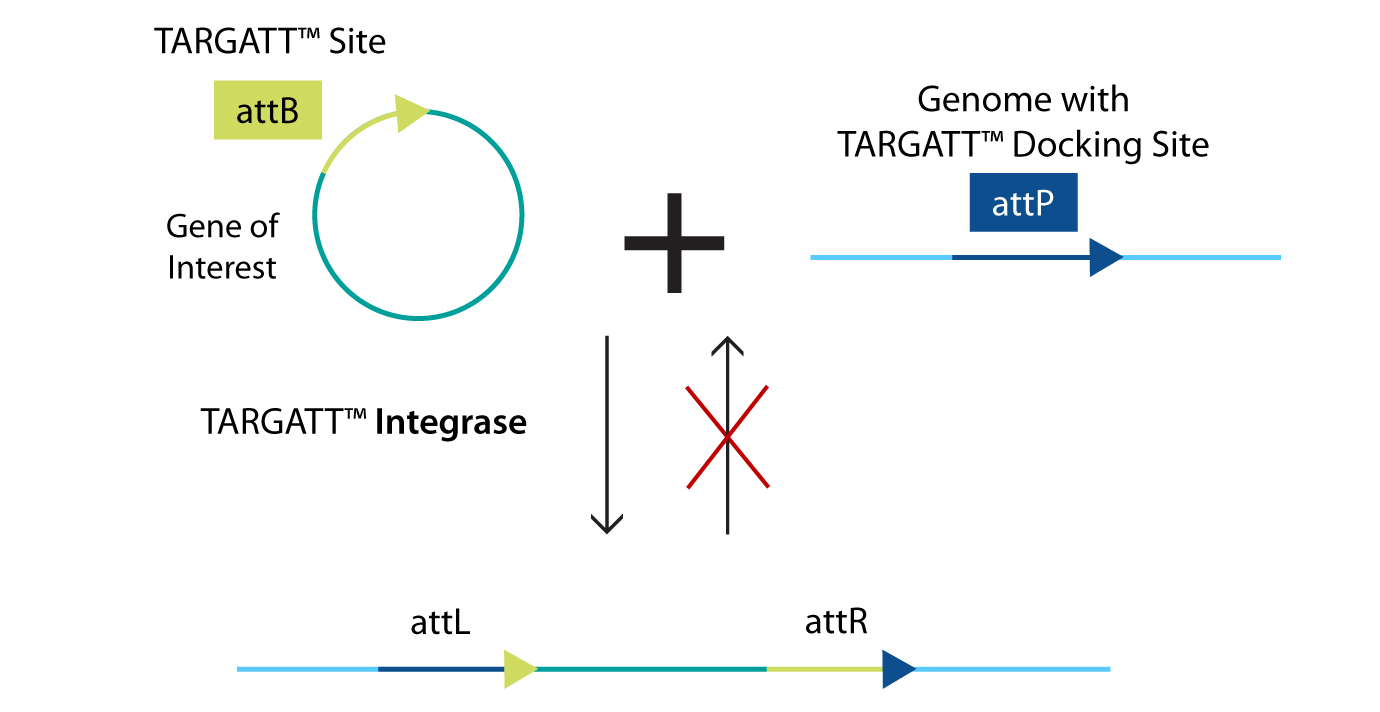
Figure 1: TARGATT™ Knock-In Strategy. TARGATT™ technology enables fast and site-specific, stable integration of large DNA fragments (up to 22 kb) into an intergenic, transcriptionally active safe harbor locus. We can engineer an "attP" integrase recognition landing pad at a safe harbor locus. Single-copy gene insertion occurs when it is used in conjunction with an “attB” containing donor plasmid and integrase expression.