Newsletter
Neuronal Disease Specific Isogenic Knockout iPSCs
Applied StemCell has engineered isogenic panels of extensively characterized, genome edited human iPSC lines that have been shown to recapitulate abnormal neuronal phenotypes seen in patient-derived iPSC.
-
Engineered iPSCs with knockout (KO) of genes associated with neurological disorders
-
Isogenic panels of iPSCs from a well-characterized control/ master iPSC line
-
Fully characterized KO iPSCs: pluripotency, differentiation, and functional phenotypes of differentiated neurons
-
Overcomes problems with tissue sourcing from diseased patients and varied genetic background which makes results hard to interpret
Our engineered hiPSC lines are physiologically relevant cell lines to model Parkinson’s disease (PD), Alzheimer’s disease, ALS, Schizophrenia, and Autism.
Neuronal Disease-Specific Isogenic Knockout Lines:
Isogenic Knock-out Lines |
Associated Disease |
Description |
|
PARK2 -/- |
PD |
Biallelic KO |
|
PARK7 -/- |
PD |
Biallelic KO |
|
PINK1 -/- |
PD |
Biallelic KO |
|
LRRK2 -/- |
PD |
Biallelic KO |
|
Park2-/-; Park7-/- |
PD |
Biallelic KO |
|
Park2-/-; Pink1-/- |
PD |
Biallelic KO |
|
APOE -/- |
Alzheimer's disease |
Biallelic KO |
|
SOD1 -/- |
ALS |
Biallelic KO |
|
DISC1 -/- |
Schizophrenia |
Biallelic KO |
|
CNTNAP2 -/- |
Autism |
Biallelic KO |
|
BDNF -/- |
CNS |
Biallelic KO |
Products and Services
Technical Details
If you need gene knockout or point mutation in specific genes in iPSCs, we provide expert custom service for iPSC- Gene Editing. We also provide custom differentiation service for the lineage of your choice.
Please inquire for details on differentiation of these genome-edited cell lines as well as custom differentiation services.
Case Studies
Case Study 1
Disease-specific Knockout iPSC Line: APOE Knockout
This iPSC line is engineered with a bi-allelic (homozygous) knockout for APOE gene (APOE-/-) that has been implicated in Alzheimer’s disease etiology. The parental iPSC line is an integration-free, normal karyotype line derived from male, CD34+ cord blood cells. The APOE knockout line can be further differentiated into an isogenic panel of neurons and glia for disease modeling and drug/ toxicity screening applications.
1. Genotyping of APOE-/- bi-allelic KO clone F4: APOE KO

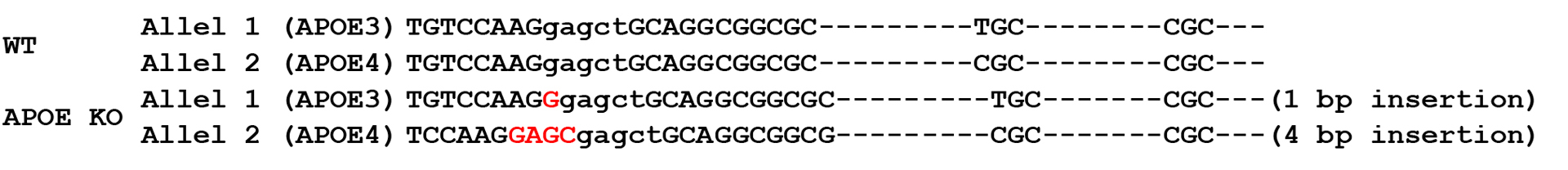
Figure 1. Sequence alignment between parental/ control iPSC line and APOE-/- clone. The homozygous knockout clone shows a 4 bp deletion in allele 1, a 3 bp deletion in allele 2, and a 4bp insertion in allele 2, as compared to wild type (WT) sequence in parental iPSC line.
2. Expression of Pluripotency Markers
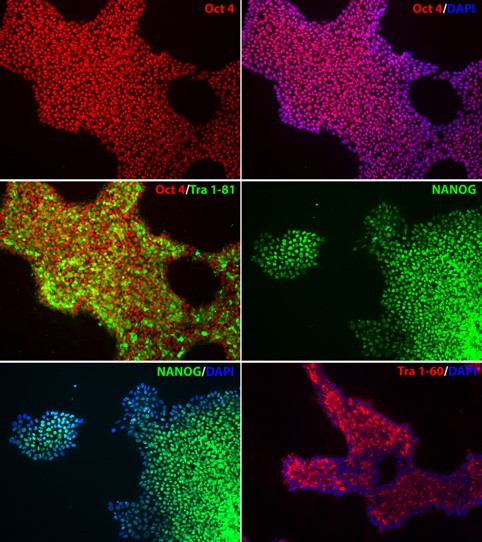
Figure 2. Expression of pluripotency markers in APOE-/- iPSC line. The homozygous knockout iPSC line, APOE-/-, expresses pluripotency markers OCT4, NANOG, TRA-1-60, and TRA-1-81, indicating pluripotency of the iPSC line after genome editing. Nucleus stained with DAPI (blue).
3. Expression of APOE in neural stem cells differentiated from APOE-/- iPSC-line
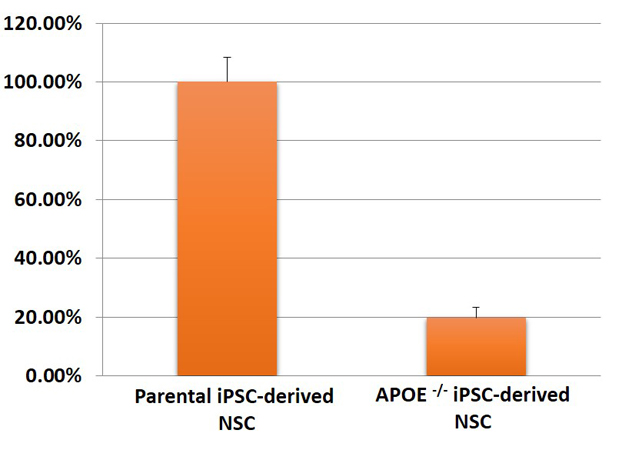
Figure 3. Expression of APOE in neural stem cells (NSC) differentiated from master/ parental iPSC line and APOE-/- iPSC line (ASE-9405) was quantified using qPCR. The APOE mRNA was significantly downregulated in the APOE-knockout NSC as compared to expression in the parental/ control iPSC line.
| WT | APOE-/- |
| 1022 | -4 |
Figure 4. APOE Expression in APOE knockout NSCs by microarray. APOE expression in the knockout NSC line was significantly lower than the wild type NSC line.
4. Differentiation of APOE -/- iPSCs into Neurons and GFAP
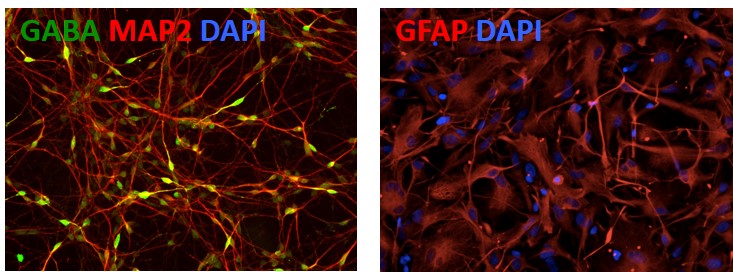
Figure 5. Differentiation of APOE-/- iPSCs into cortical neurons and astrocytes. The APOE-/- line can be differentiated into cortical neurons (GABA; green) and astrocytes (GFAP; red) to model Alzheimer's disease. Other markers: Neuronal marker (MAP2; red) and nucleus staining (DAPI; blue).
Application Notes
Advances in neurological disease treatment have suffered because of a lack of physiologically relevant and reliable disease models that recapitulates the disease phenotypes seen in patients. Predominantly used rodent models and patient-derived primary cells for studying underlying cellular and genetic pathology and drug discovery have frequently proven to be unreliable when translating results to human studies. The discovery of induced pluripotent stem cell (iPSC) and CRISPR gene editing technologies has launched a new era in neuroscience research and drug discovery. While, disease modeling using iPSCs derived from patients has been shown to recapitulate cellular pathology seen in patient tissues, the varied genetic background of patient-derived cells poses a significant challenge to analyze and interpret results. Genetically modified iPSCs can overcome the problems associated with tissue sourcing and genetic backgrounds. CRISPR technology allows for accurately modeling a variety of disease mutations in the same genetic background (isogenic) as the parent cell line. The use of matched isogenic control provides a reliable control for interpreting experimental data. Such genetically modified iPSCs have shown to be a biorelevant model of human physiology and diseases, are easy to source, have a great potential in personalized regenerative medicine.
Applications:
- Physiologically relevant cell lines to model Parkinson’s disease, Alzheimer’s disease, ALS, Schizophrenia, and Autism.
- Predictive models to study neurogenetic mechanisms
- Suitable for high-throughput drug screening of neuroprotective and neurotoxicological compounds using differentiated neuronal lineages.




