Newsletter
TARGATT™ CHO and HEK293 Cells For Antibody Library Construction
TARGATT™ technology enables faster and more efficient site-specific integration of large DNA fragments in cell lines. Our proprietary technology offers an ideal platform for generating stable cell line libraries for mammalian cell display-mediated antibody engineering, protein evolution screening, mammalian two-hybrid (M2H) screens, and more. It allows only a 1:1 variant-to-cell ratio with a uniform and consistent expression of the gene/ protein for efficient screening.
At ASC, we integrated TARGATT™ site-specific knock-in technology into CHO and HEK293 cells. The TARGATT™ Master CHO and HEK293 Cell Lines enable single-copy insertion at a safe-harbor locus (H11, ASC2). High integration efficiencies and medium to high levels of protein expression have been observed.
The TARGATT™ master cell lines are ideal for antibody library construction. Our experts have successfully completed several custom TARGATT™ HEK293- and CHO-based library construction projects which have enabled the optimization of our off-the-shelf TARGATT™ HEK293 and CHO library screening kits (Drug selection and FACS kits available). These high-quality kits allow you to build your library in your own lab! Explore our products below or contact us today to schedule your free consultation.
ASC TARGATT™ Advantages:
- Single copy knockin: 1 cell, 1 docking site, 1 inserted transgene
- Site-specific knockin into a high expression, safe harbor locus (H11, ASC2)
- High efficiency integration (HEK293: >40% without, >90% with drug selection; CHO: ~18% without, >90% with drug selection)
- Large cell library construction in HEK293/ CHO cells
- Cost-effective: no virus packaging time and resources
- BSL1 compatible
- Fully customizable library construction service
- Four ready-to-use TARGATT™ antibody library screening kits for you to build your own library
Licensing and Evaluation Programs Available!
Products and Services
Technical Details
TARGATT™ Library Screen Master Cell Lines: HEK293 & CHO
What do current library screening systems lack?
Traditionally, library screening is completed using bacteria or yeast phage display. Bacteria allow for the creation of large libraries in a short period of time while yeast provides a eukaryotic environment. These systems may be cost-effective, but they lack posttranslational modifications that mammalian cells permit.
Mammalian cells also offer a human-like environment, but the currently available systems are slow and laborious to work with at a high cost. The available mammalian library systems for screening may provide an environment closer to the human system, but the coverage they allow is very low compared to bacteria and yeast.
Applied StemCell’s Solution
Our goal was to engineer the TARGATT™ system into HEK293 & CHO cells in order to:
- Develop a mammalian display system with a higher efficiency
- Provide a mammalian display system that can reach E. coli and
yeast library sizes
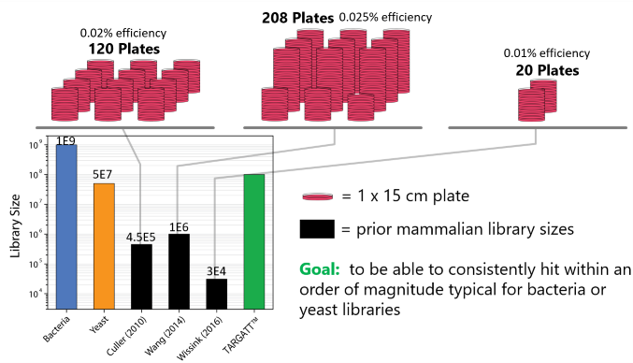
Figure 1: Comparison of the currently available library screening systems.
TARGATT™ Screen Master Cell Lines
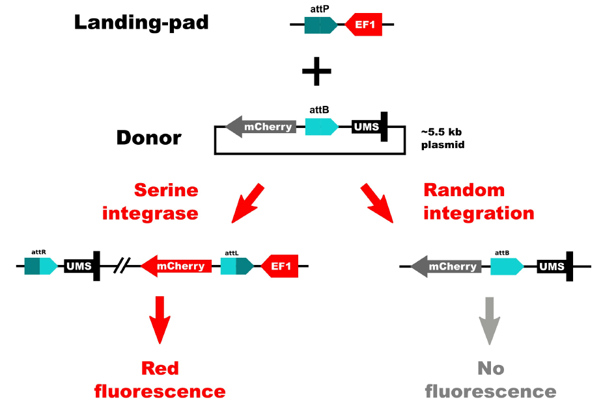
The TARGATT™ Screen Master Cell Lines were engineered using a split-cassette selection/screen system. This allows us to obtain clean results with little background. We separated the promoter and the transgene. The promoter was inserted in the chromosome at a safe-harbor locus, and the transgene is carried by the donor plasmid. This system only allows expression of the insert if there is a site-specific gene insertion at the safe-harbor locus that contains the promoter. If random integration were to occur, the gene would not have a promoter and therefore would not be expressed.
Applied StemCell integrated this system into HEK293 and CHO cells. Both cell lines have reported high integration efficiencies and medium to high levels of protein expression.
Figure 2: Schematic of the split-cassette selection/screen system.
TARGATT™ Mammalian Display
To address the current library screening and size issues, Applied StemCell is using its TARGATT™ gene editing technology to develop a mammalian display system that can consistently hit within an order of magnitude typical for bacteria and yeast. When comparing the TARGATT™ system for mammalian display to other available systems, it is clear that the TARGATT™ system offers unique features including site-specific and single-copy gene insertion.
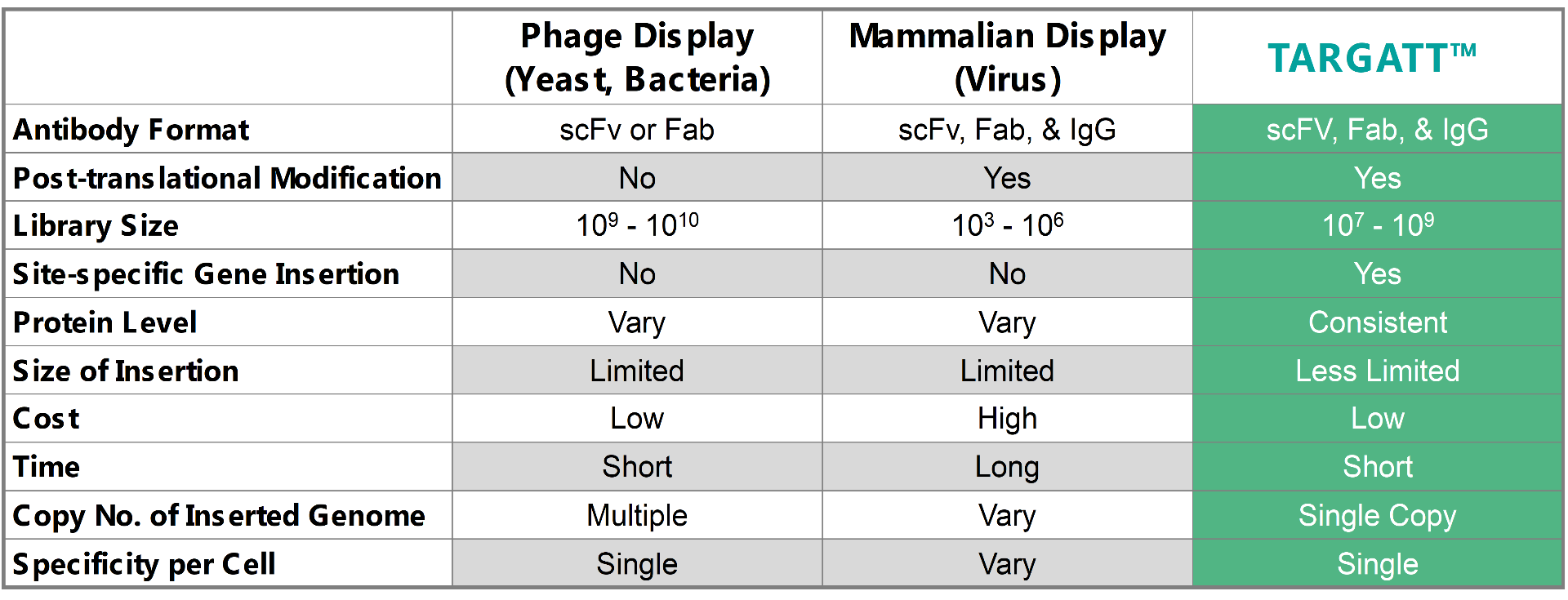
Table 1: A comparison of the TARGATT™ mammalian display with available alternative display systems.
eBook: TARGATT™ Technology For Antibody Discovery and Screening (March 2021)
*Featured in Informa Connect's eBook: Antibody Discovery, Selection & Screening.
Support Materials
eBook:
TARGATT™ Technology For Antibody Discovery and Screening
(March 2021)
*Featured in Informa Connect's eBook: Antibody Discovery, Selection & Screening.
Webinar:
![]() TARGATT™ Unique Technology for Antibody Engineering & Screening
TARGATT™ Unique Technology for Antibody Engineering & Screening
(February 2021)
Poster and Poster Presentation:
![]() Construction of a Naïve Human Immunoglobin Mammalian Cell Library Using TARGATT™ Technology
Construction of a Naïve Human Immunoglobin Mammalian Cell Library Using TARGATT™ Technology
(December 2020)
![]() Construction of a Naïve Human Immunoglobin Mammalian Cell Library Using TARGATT™ Technology
Construction of a Naïve Human Immunoglobin Mammalian Cell Library Using TARGATT™ Technology
(December 2020)
Publications
TARGATT™ Master Cell Lines
- Chi, X., Zheng, Q., Jiang, R., Chen-Tsai, R. Y., & Kong, L. J. (2019). A system for site-specific integration of transgenes in mammalian cells. PLOS ONE, 14(7), e0219842.
Transgenic Mouse Book Chapters
- Chen-Tsai, R. Y. (2020). Integrase-Mediated Targeted Transgenics Through Pronuclear Microinjection. In Transgenic Mouse (pp. 35-46). Humana, New York, NY.
- Chen-Tsai, R. Y. (2019). Using TARGATT™ Technology to Generate Site-Specific Transgenic Mice. In Microinjection (pp. 71-86). Humana Press, New York, NY.
Description of the technology
- Zhu, F., Gamboa, M., Farruggio, A. P., Hippenmeyer, S., Tasic, B., Schüle, B., … Calos, M. P. (2014). DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Research, 42(5), e34. http://doi.org/10.1093/nar/gkt1290.
- Tasic, B., Hippenmeyer, S., Wang, C., Gamboa, M., Zong, H., Chen-Tsai, Y., & Luo, L. (2011). Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proceedings of the National Academy of Sciences of the United States of America, 108(19), 7902–7907. http://doi.org/10.1073/pnas.1019507108.
Commentary, comparison with other transgenic methods
- Rossant, J., Nutter, L. M., & Gertsenstein, M. (2011). Engineering the embryo. Proceedings of the National Academy of Sciences, 108(19), 7659-7660.
Tet inducible mice generated by TARGATT™
- Fan, X., Petitt, M., Gamboa, M., Huang, M., Dhal, S., Druzin, M. L., … Nayak, N. R. (2012). Transient, Inducible, Placenta-Specific Gene Expression in Mice. Endocrinology, 153(11), 5637–5644. http://doi.org/10.1210/en.2012-1556.
Advantage of Hipp11 (H11) locus
- Hippenmeyer, S., Youn, Y. H., Moon, H. M., Miyamichi, K., Zong, H., Wynshaw-Boris, A., & Luo, L. (2010). Genetic Mosaic Dissection of Lis1 and Ndel1 in Neuronal Migration. Neuron, 68(4), 695–709. http://doi.org/10.1016/j.neuron.2010.09.027.
Applications for mice generated by TARGATT™ (and cited/published articles
- Lindtner, S., Catta-Preta, R., Tian, H., Su-Feher, L., Price, J. D., Dickel, D. E., ... & Pennacchio, L. A. (2019). Genomic Resolution of DLX-Orchestrated Transcriptional Circuits Driving Development of Forebrain GABAergic Neurons. Cell reports, 28(8), 2048-2063.
- Wang, T. A., Teo, C. F., Åkerblom, M., Chen, C., Tynan-La Fontaine, M., Greiner, V. J., ... & Jan, L. Y. (2019). Thermoregulation via Temperature-Dependent PGD2 Production in Mouse Preoptic Area. Neuron, 103(2), 309-322.
- Clarke, B. A., Majumder, S., Zhu, H., Lee, Y. T., Kono, M., Li, C., ... & Byrnes, C. (2019). The Ormdl genes regulate the sphingolipid synthesis pathway to ensure proper myelination and neurologic function in mice. eLife, 8.
- Carlson, H. L., & Stadler, H. S. (2019). Development and functional characterization of a lncRNA‐HIT conditional loss of function allele. genesis, e23351.
- Chande, S., Ho, B., Fetene, J., & Bergwitz, C. (2019). Transgenic mouse model for conditional expression of influenza hemagglutinin-tagged human SLC20A1/PIT1. PloS one, 14(10), e0223052. doi:10.1371/journal.pone.0223052
- Hu, Q., Ye, Y., Chan, L. C., Li, Y., Liang, K., Lin, A., ... & Pan, Y. (2019). Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nature immunology, 1.
- Matharu, N., Rattanasopha, S., Tamura, S., Maliskova, L., Wang, Y., Bernard, A., ... & Ahituv, N. (2018). CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science, eaau0629.
- Barrett, R. D., Laurent, S., Mallarino, R., Pfeifer, S. P., Xu, C. C., Foll, M., ... & Hoekstra, H. E. (2018). The fitness consequences of genetic variation in wild populations of mice. bioRxiv, 383240.
- Ibrahim, L. A., Huang, J. J., Wang, S. Z., Kim, Y. J., Li, I., & Huizhong, W. (2018). Sparse Labeling and Neural Tracing in Brain Circuits by STARS Strategy: Revealing Morphological Development of Type II Spiral Ganglion Neurons. Cerebral Cortex, 1-14.
- Kumar, A., Dhar, S., Campanelli, G., Butt, N. A., Schallheim, J. M., Gomez, C. R., & Levenson, A. S. (2018). MTA 1 drives malignant progression and bone metastasis in prostate cancer. Molecular oncology.
- Jang, Y., Broun, A., Wang, C., Park, Y. K., Zhuang, L., Lee, J. E., ... & Ge, K. (2018). H3. 3K4M destabilizes enhancer H3K4 methyltransferases MLL3/MLL4 and impairs adipose tissue development. Nucleic acids research. https://doi.org/10.1093/nar/gky982
- Tang, Y., Kwon, H., Neel, B. A., Kasher-Meron, M., Pessin, J., Yamada, E., & Pessin, J. E. (2018). The fructose-2, 6-bisphosphatase TIGAR suppresses NF-κB signaling by directly inhibiting the linear ubiquitin assembly complex LUBAC. Journal of Biological Chemistry, jbc-RA118.
- Chen, M., Geoffroy, C. G., Meves, J. M., Narang, A., Li, Y., Nguyen, M. T., ... & Elzière, L. (2018). Leucine Zipper-Bearing Kinase Is a Critical Regulator of Astrocyte Reactivity in the Adult Mammalian CNS. Cell Reports, 22(13), 3587-3597
- Kido, T., Sun, Z., & Lau, Y.-F. C. (2017). Aberrant activation of the human sex-determining gene in early embryonic development results in postnatal growth retardation and lethality in mice. Scientific Reports, 7, 4113. http://doi.org/10.1038/s41598-017-04117-6.
- Nouri, N., & Awatramani, R. (2017). A novel floor plate boundary defined by adjacent En1 and Dbx1 microdomains distinguishes midbrain dopamine and hypothalamic neurons. Development, 144(5), 916-927.
- Li, K., Wang, F., Cao, W. B., Lv, X. X., Hua, F., Cui, B., ... & Yu, J. M. (2017). TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of p53-Mediated Senescence. Cancer Cell, 31(5), 697-710.
- Jiang, T., Kindt, K., & Wu, D. K. (2017). Transcription factor Emx2 controls stereociliary bundle orientation of sensory hair cells. eLife, 6, e23661.
- Booze, M. L., Hansen, J. M., & Vitiello, P. F. (2016). A Novel Mouse Model for the Identification of Thioredoxin-1 Protein Interactions. Free Radical Biology & Medicine, 99, 533–543. http://doi.org/10.1016/j.freeradbiomed.2016.09.013.
- Feng, D., Dai, S., Liu, F., Ohtake, Y., Zhou, Z., Wang, H., ... & Hayat, U. (2016). Cre-inducible human CD59 mediates rapid cell ablation after intermedilysin administration. The Journal of clinical investigation, 126(6), 2321-2333.
- Sun, N., Yun, J., Liu, J., Malide, D., Liu, C., Rovira, I. I., … Finkel, T. (2015). Measuring in vivo mitophagy. Molecular Cell, 60(4), 685–696. http://doi.org/10.1016/j.molcel.2015.10.009.
- Devine, W. P., Wythe, J. D., George, M., Koshiba-Takeuchi, K., & Bruneau, B. G. (2014). Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife, 3, e03848. http://doi.org/10.7554/eLife.03848.
- Fogg, P. C. M., Colloms, S., Rosser, S., Stark, M., & Smith, M. C. M. (2014). New Applications for Phage Integrases. Journal of Molecular Biology, 426(15), 2703–2716. http://doi.org/10.1016/j.jmb.2014.05.014.
- Chen-Tsai, R. Y., Jiang, R., Zhuang, L., Wu, J., Li, L., & Wu, J. (2014). Genome editing and animal models. Chinese science bulletin, 59(1), 1-6.
- Park, K.-E., Park, C.-H., Powell, A., Martin, J., Donovan, D. M., & Telugu, B. P. (2016). Targeted Gene Knockin in Porcine Somatic Cells Using CRISPR/Cas Ribonucleoproteins. International Journal of Molecular Sciences, 17(6), 810. http://doi.org/10.3390/ijms17060810.
- Guenther, C. A., Tasic, B., Luo, L., Bedell, M. A., & Kingsley, D. M. (2014). A molecular basis for classic blond hair color in Europeans. Nature Genetics, 46(7), 748–752. http://doi.org/10.1038/ng.2991.
- Villamizar, C. A. (2014). Characterization of the vascular pathology in the acta2 r258c mouse model and cerebrovascular characterization of the acta2 null mouse. UT GSBS Dissertations and These (Open Access). Paper 508 (2014)
FAQs
Can we in this case fuse the signal sequence directly to the C-terminus of the P2A tag or do we need to attach a linker sequence?
Does the vector cassette (mCHERRY-P2A-LacZ) has PaqC1 cleavage sites (CACCTGC)?
You can also request the construction of a library, but can you give us an estimate of the price?
There are four types of kits for antibody library screening, but how do you plan to use them properly? Which one is better suited for building as large a library as possible?
Which TARGATT™ CHO Kit should we use if we are not isolating single antibodies (eg. the best binders)?
What is the difference between AST-1400, AST-1405, AST-1409, and AST-1410?
Which TARGATT™ CHO Kit should I use?
What is the difference between the AST-1306/AST-1307 antibody screening kits and the AST-1305 knockin kit?
Why should I use ASC's HEK293/CHO TARGATT system rather than phage-display or other display methods?
Can I use this system in my own cell line?
Can you tell us a bit more about the screening process, does it require clonal selection?
Could you describe the business model to access your technology?
You can do up to 10e9, but is it possible to provide a protocol for building a large-scale library?
Is the library constructing plasmid available for us to make our own library by ourselves?
What can the TARGATT™ library system be used for?
Why is TARGATT™ better?
How long does it take to obtain a pool? Could this be used for high-throughput Ab production?
AST-1307: Is there a marker for detection of integration efficiency?
AST-1306 & AST-1307: Is additional integrase available?
AST-1306 & AST-1307: How many screenings can we do with these kits?
AST-1306 & AST-1307: I want to test this. What is the next step?
With more copies will I have a higher yield?
AST-1306 & AST-1307: Are you able to make a construct library for the GOI that I have?
Why do we need to use Golden Gate Assembly for the cloning?
AST-1409 and 1410: I am interested in the kit, what information can I obtain in order to insert my GOI?
AST-3080 and 3081: Do you disclose the termination site?
AST-3080 and 3081: Do you disclose the promoter, and its sequence?
Why do we need to use these special 10G SUPREME SOLO cells for transformation?
AST-1409: A plasmid was transfected in by electroporation or lipofectamine?
AST-1409: A plasmid was introduced by lentiviral or other retrovirus methods?
AST-1409: The cell line was modified using CRISPR/Cas9 engineering?
Regarding the TARGATT™ 41 attB-mCherry-P2A-LacZ (library) Cloning Plasmid: After cloning the gene/s of interest into this plasmid using the NEB kit, the protocol recommends transforming using E. cloni® 10G SUPREME SOLOs electrocompetent cells. How would y
In the treatise (PLOS ONE, 14 (7), e0219842.), The GFP positive rate before GCV selection is 12.2% even in HEK, but it is ~ 40% described on the website. Is this due to the difference between the paper and the system?
In the one in the paper, it seems that PhiC is also incorporated in the Host Cell, but is it the type that Kit also introduces CMV-integrase plasmid at the same time? If possible, could you please tell me why you are changing?
Is there a recommended protocol for displaying antibodies? It will be helpful if you have information on how to express Hch / Lch and what to anchor.
Information on gene transfer in CHO is shown in Neon, but is there any information in other devices (NEPA, Nucleofactor2b, etc.)?
We recovered the AST-1408-1 cells sent with the AST-1409 kit using a different media (CD FortiCHO™), and the cells did not recover well (low viability). Could I please get some guidance on this issue? I think the issue was the seeding density.