Newsletter
CRISPR Knock-in & Point Mutation iPS Cell Generation
Save your Time, Money & Effort with our CRISPR-iPSC high throughput, automated platform for knock-in and point mutation iPS cell engineering. Applied StemCell (ASC) can produce viable, pluripotent iPSCs with any genetic modification you require, including single or double gene knockin, SNV corrections, and more. Our custom iPSC knock-in and point mutation services allow you to choose between 2 high-quality heterozygous or homozygous clones, or you can purchase additional clones for your basic research, disease modeling, cell therapy, or isogenic cell line development projects.
- High-throughput automation for high volume projects and lower cost
- High-efficiency
- Insert gene of interest or reporter genes in iPSCs
- Point Mutation in iPSCs or correct mutations in WT
- Standard and complex genetic modifications available: point mutations, SNV corrections, reporter/tag knockin, double gene knockin, and more
- Heterozygous or homozygous clones
- Most updated CRISPR designs
- Faster timelines than other providers
- GMP iPSC Gene Editing Available >> Learn More
ASC also supplies CRISPR-editable and differentiation-proven control iPSC lines. We even offer downstream differentiation and assay development services. If you would like to learn more about how you can make the most of our CRISPR-iPSC service platform, schedule a free consultation today.
Workflow for iPSC Knock-in or Point Mutation

Products and Services
Case Studies
Case Study #1: GFP Reporter Knock-in in iPSCs
Goal: The purpose of this project was to genetically introduce a GFP reporter tag into control human iPSCs at a specified locus "A".
Two gRNA candidates were selected based on the proximity to the knock-in sites and off-target profiles, and functionally validated in a model cell line. The gRNAs that produced the desired NHEJ frequencies were then used for the transfection into the control human iPSC line. Single cell colonies were screened by genotyping
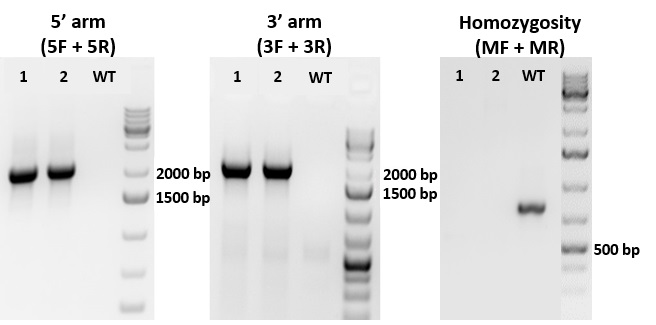
Figure 1. PCR genotyping screening of GFP knock-in at locus "A". Three sets of PCR primers for each knock-in line were designed to amplify PCR fragments flanking left homology region (5 arm), right homology region (3 arm), and reporter gene insertion region (M) (to identify homozygous clones). Both Clone#1 and 2 were homozygous as attested by absence of PCR band (when compared to WT) and further by Sanger Sequencing (not shown).
Case Study #2: Point Mutation Correction of a Mutant Allele in a Human Induced Pluripotent Stem Cell Line
Goal: The goal of this project was to correct a point mutation (single nucleotide polymorphism; SNP) found in a mutant allele of the gene-of-interest in a patient-derived iPSC line.
The point mutation was corrected by co-transfection of CRISPR reagents: Cas9, validated gRNA, and a single stranded oligodeoxynucleotide donor (ssODN) into the iPSCs. The gRNAs were designed based on the proximity to the mutation seen in the mutant allele and functionally validated for optimal NHEJ frequency. The ssODN was designed to replace the mutation with wildtype sequence using homology directed repair (HDR). Off-target analysis was also performed for each gRNA (not shown). After transfection, single clones were isolated and genotyped to confirm desired mutation correction. One corrected clone was identified and confirmed by sequencing (Figure 2A and 2B).
A. 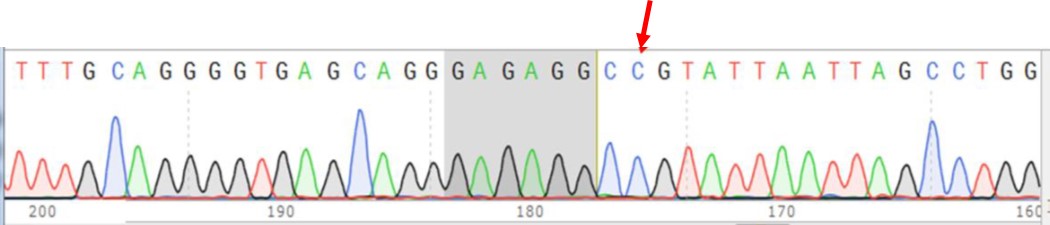

B.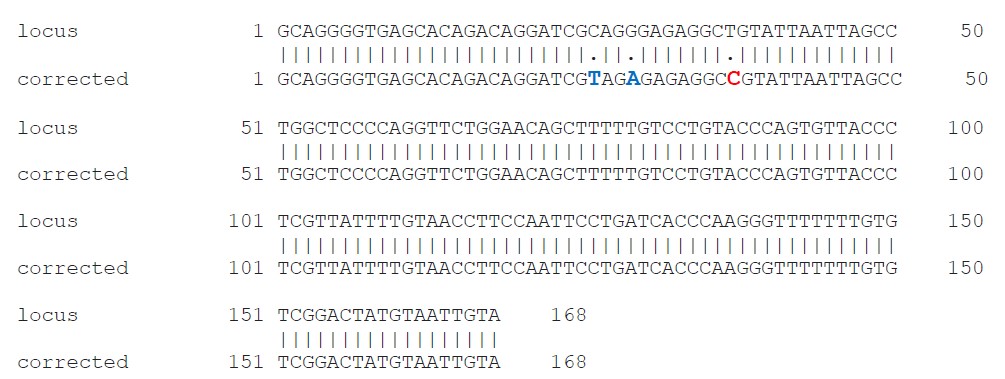

Figure 2. (A) Sequencing chromatogram of corrected clone (CTG > CCG). Red arrow indicates the nucleotide correction from a CTG to CCG. (B) Sequence alignment of corrected clone (bottom) against the parental SNP sequence (top). Two silent mutations were introduced by donor ssODN (CGC > CGT, GGG to GAG).
Case Study #3: Point Mutation Correction in a Patient-Derived iPSC
Goal: To correct a point mutation (AGC > GGC) in a gene associated with neurological disorders in a patient derived iPSC line and generate a homozygous wildtype line.
1. Patient-derived iPSCs were obtained from the patient and the cell line validated. The sequence at the desired locus (neurological gene) was confirmed to be a heterozygous mutation (AGC/GGC).
Figure 1. Schematic representation of the workflow undertaken for the project.
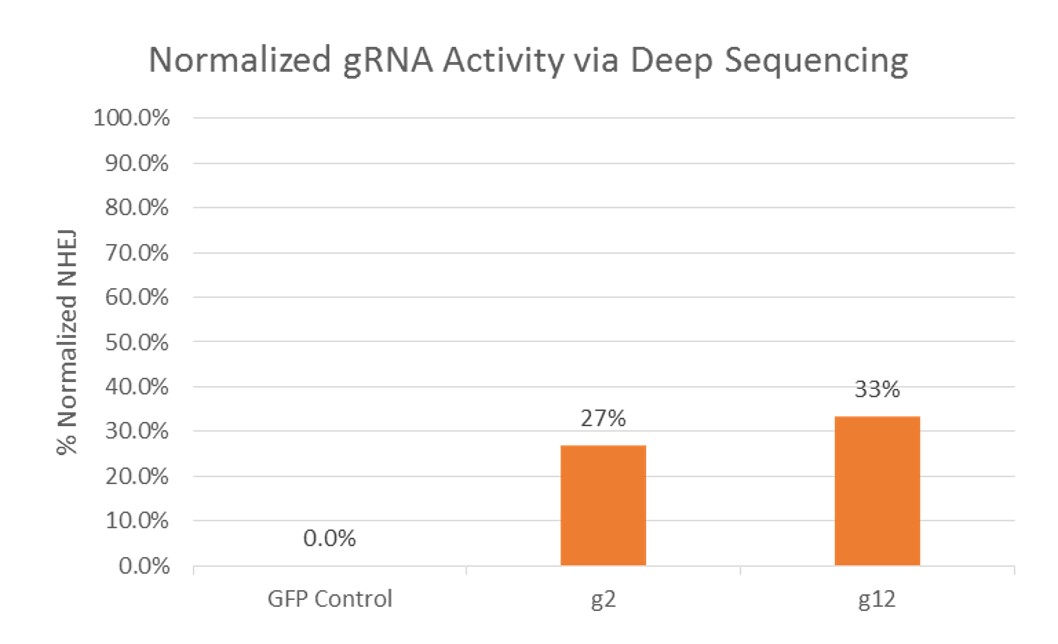
2. After cell line validation and confirming presence of mutation in the desired gene, two gRNAs were designed and validated by mismatch detection assay in K562 cells and frequency of NHEJ events as a result of the gRNA-Cas9 complex and DSB was quantified using next generation/ deep sequencing (NGS).
Figure 2. gRNA Activity via NGS, Normalized by NHEJ frequency resulting from control (GFP) transfection. Results showed that NHEJ frequency mediated by gRNA g1 and g2 were 27% and 33%, respectively. The gRNA g2 was selected for transfection.
3. The Cas/gRNA and donor vectors were transfected into the hiPSCs. After a transient puromycin selection, single cell colonies were isolated and expanded. Individual colonies were then genotyped by PCR and sequencing. Positive clones were expanded, and the genotypes were further confirmed by sequencing.
A. 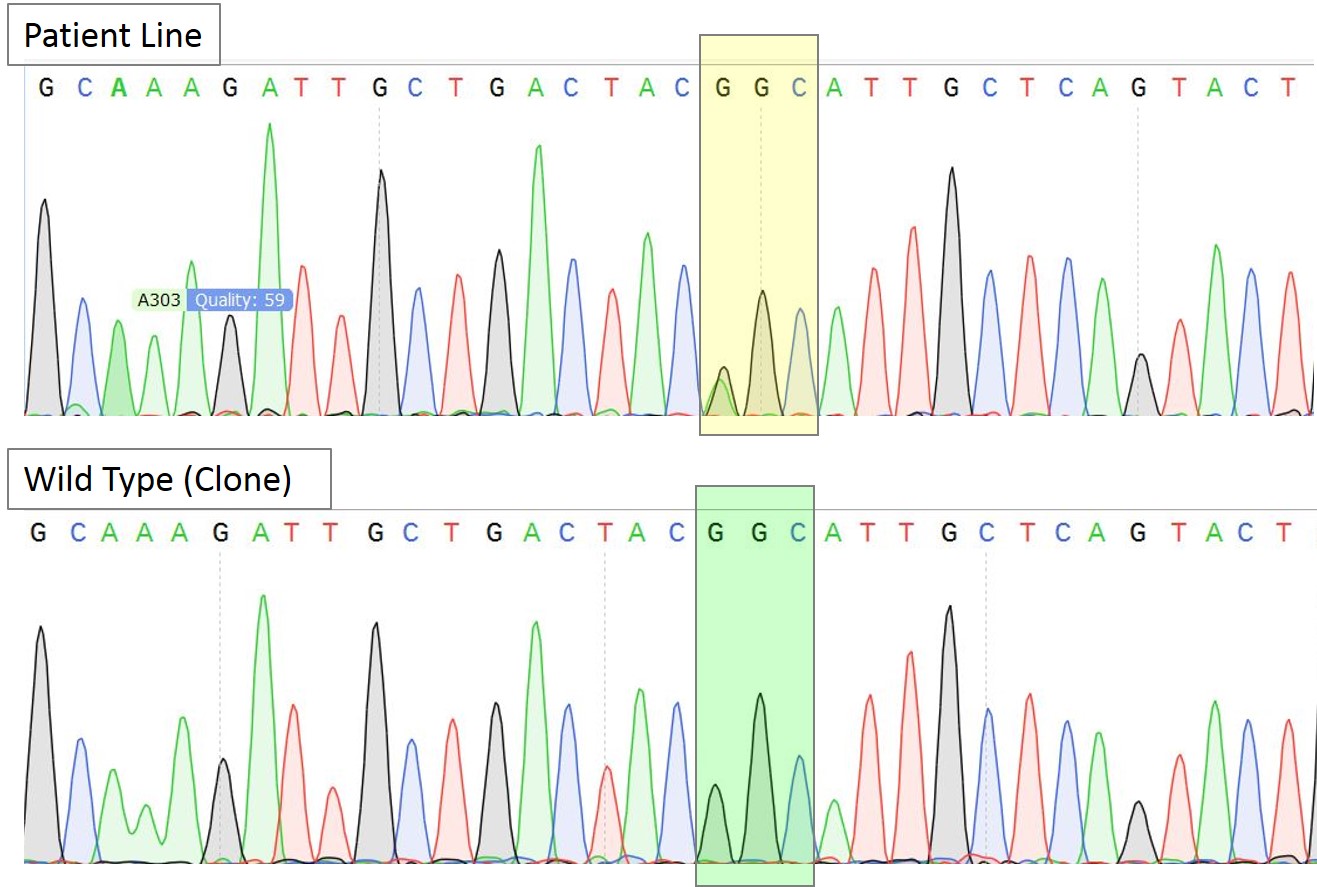
Figure 3A. Sequence chromatogram of the patient line with heterozygous mutation AGC/GGC and a representative homozygous wildtype clone. The yellow box highlights the heterozygous mutation while the green box highlights the correction to homozygous wild type sequence (GGC) in the selected clone.
B. 
Figure 3B. Sequence alignment of representative homozygous clone (WT) and the mutation (the patient line). A silent mutation (GCA; yellow) was introduced in the donor sequence to prevent repeated recognition and cutting by the Cas9/gRNA complex at the modification site. The mutation correction is highlighted in green.
Support Materials