Newsletter
CRISPR Humanized Mouse Model Generation Service
At ASC, we utilize CRISPR/Cas9 technology to produce genetically modified humanized mouse models. Our experts can design your ideal project to knock in or replace your specific mouse gene with a human gene, genomic sequence, or regulatory element. We can even design humanized models with conditional gene activation. These animal models are ideal for drug development, efficacy and safety studies, and more.
- Most up-to-date CRISPR design strategies and protocols
- 100% target-site cutting efficiency using validated gRNA
- >99% success rate in generating final mouse models
- A wide variety of genome modifications are available
- F1 breeding to confirm germline transmission
- Electrophysiology and behavioral assessments for your custom mouse models
- Fast turnaround
- Free consultation
ASC also supplies an assortment of well-characterized, off-the-shelf humanized mouse models. If you would like to learn, contact us today to set up your free consultation.
Products and Services
Technical Details
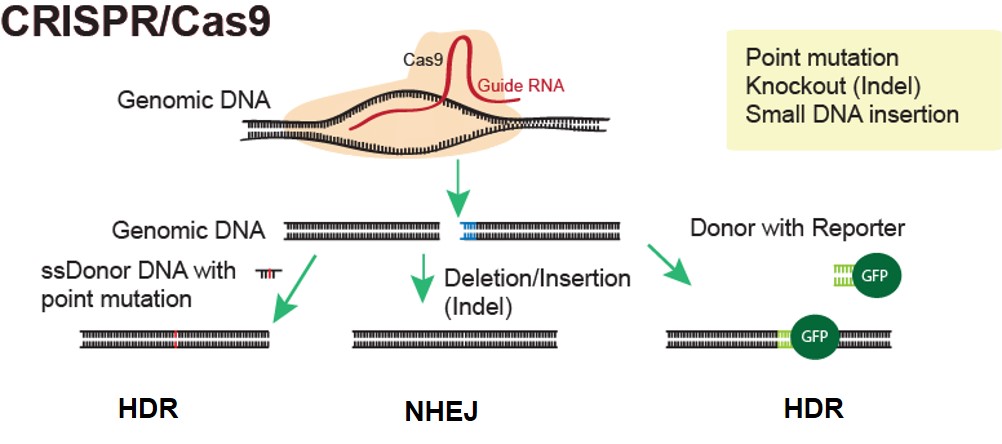
|
Service |
Time |
Deliverables |
|
1. Targeting DNA Vector Creation gRNA Design and Construction (2-4 gRNAs) |
|
A Report on Cloning and Validation |
|
2. CRISPR DNA Pronuclear Microinjection |
1-2 months |
|
|
3. Animal Care, Housing, and Genotyping Pups generated and genotyping showing the proof of precise gene insertion |
2-3 months |
At least 1 Founder |
Inquire about our CRISPR Mouse Model Generation Service: Knockout, Knock-in, Point Mutation
Case Studies
Case Study 1
CRISPR Knock-in Model - Generation of site-specific 2 kb large fragment knock-in mouse using CRISPR/Cas9
Goal: To insert a 2 kb large fragment DNA (gene of interest) at “a specified locus” in the mouse genome using CRISPR/Cas9.
The project was designed using a well-optimized protocol to generate the transgenic mice: (1) Cas9 mRNA and gRNA were produced by in vitro transcription; (2) donor vector was constructed by in-fusion method: the plasmid contained 1 kb long 5’ and 1 kb long 3’ homologous arms flanking the gene of interest (2kb); (3) the mixture of Cas9 mRNA, gRNA and donor vector was microinjected into fertilized eggs of C57BL/6j mouse background.
Using a panel of genotyping primer pairs, three out of 31 pups born after microinjection (#15, 19, and 26) were identified as founders (F0), with the gene of interest inserted at the desired locus.
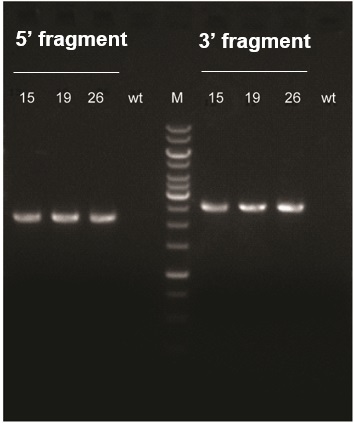
Figure 1: Agarose gel electrophoresis of PCR results in F0 mice (#15, 19, and 26) with site-specific gene knock-in. The left part of the gel shows the 5’ junction fragment (2,191 bp), and the right part of the gel shows the 3’ junction fragment (2,557 bp). [wt: wildtype control; M: 1 kb DNA ladder].
Case Study 2
CRISPR Knock-in Model - Site-specific knock-in of a 27 bp tag in mice using CRISPR/Cas9
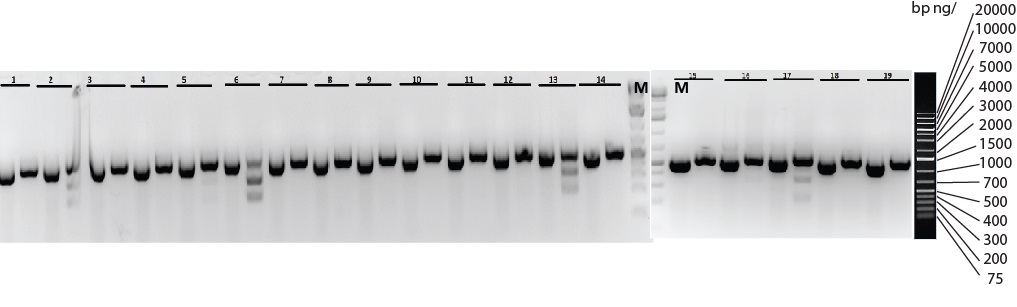
Figure: Five (#2, 5, 6 13 and 17) out of nineteen pups, 26%, were genotyped by restriction enzyme digest and sequencing to confirm the correct knock-in on one of the alleles (heterozygotes). A sequence with the desired knock-in generated a different cutting pattern compared to its wild-type counterpart.
Application Notes
Factors affecting genome editing in mouse models using CRISPR/Cas9
Three major factors influence the efficiency of CRISPR/Cas9 in mouse models: the concentrations of Cas9 mRNA, gRNA, and donor DNA. Each of these factors is interdependent which in turn affects three other parameters: the nuclease-mediated cutting rate, gene modification efficiency, and birth rate of the pups.
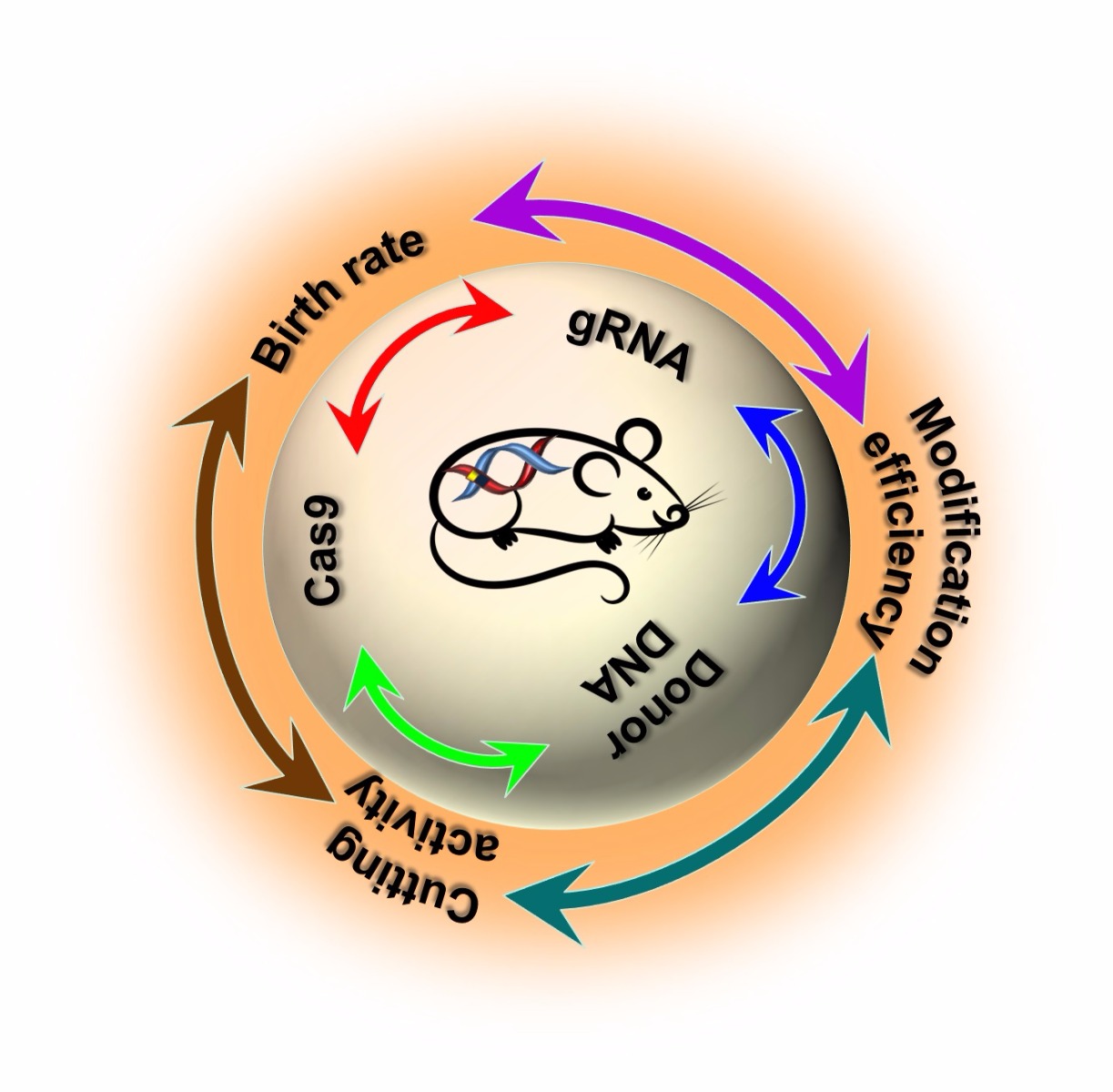
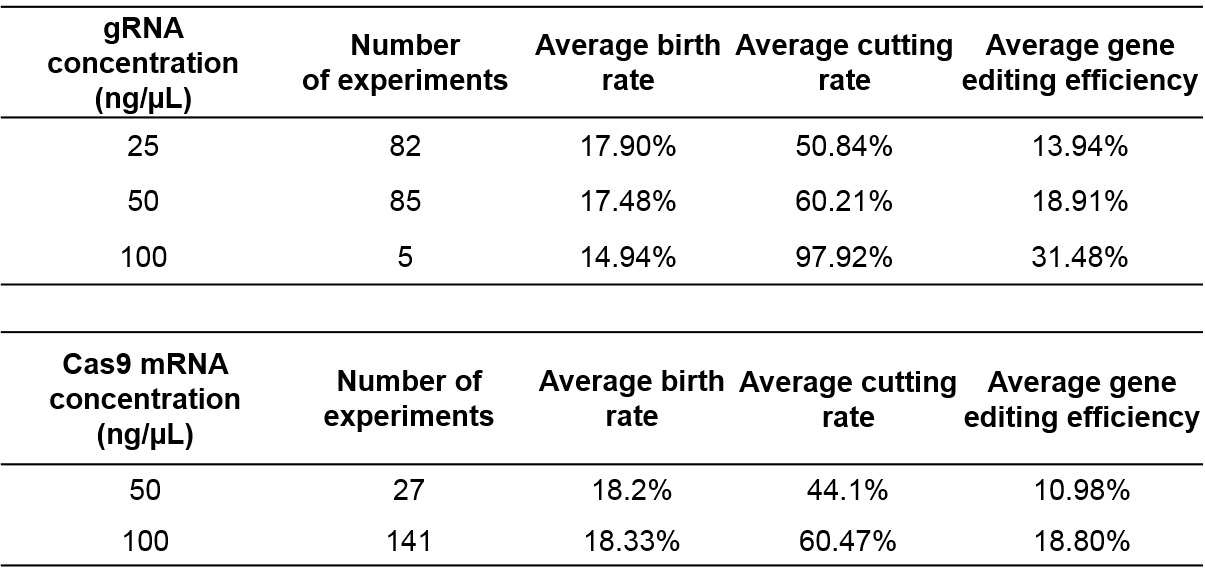
Increasing gRNA and Cas9 concentration improves cutting/editing efficiency but affects the birth rate
With increasing gRNA and Cas9 mRNA concentrations, the cutting efficiency increases to almost 97%, but the birth rate decreases. Therefore, a balance between gRNA and Cas9 concentrations, and the birth rate of mice is required for maintaining a reasonable number of embryos microinjected and in turn to optimize birthrate and number of founders born.
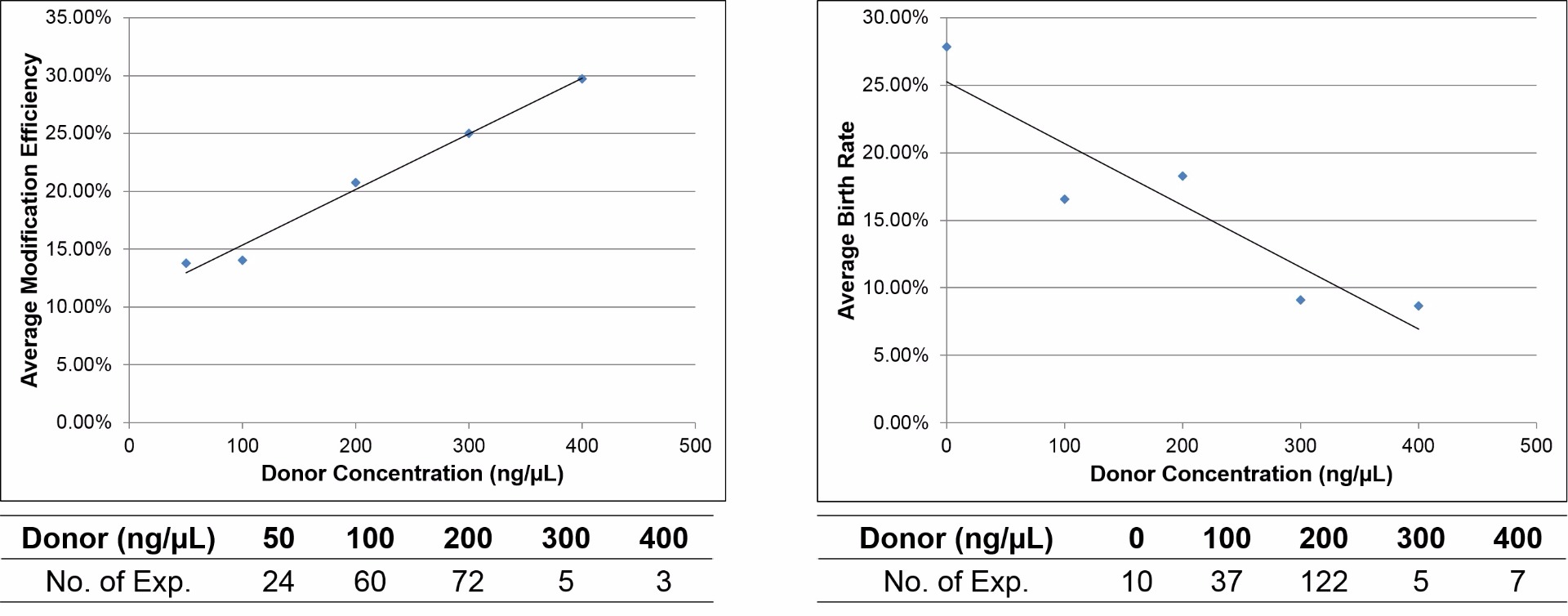
Optimal donor concentration is also necessary to maintain a balance between gene editing efficiency and birth rate
With increasing donor DNA concentration, the gene editing efficiency of the CRISPR system increases but the birth rate decreases. An optimal donor concentration of 200 ng/µl is recommended for microinjection in mouse embryos.
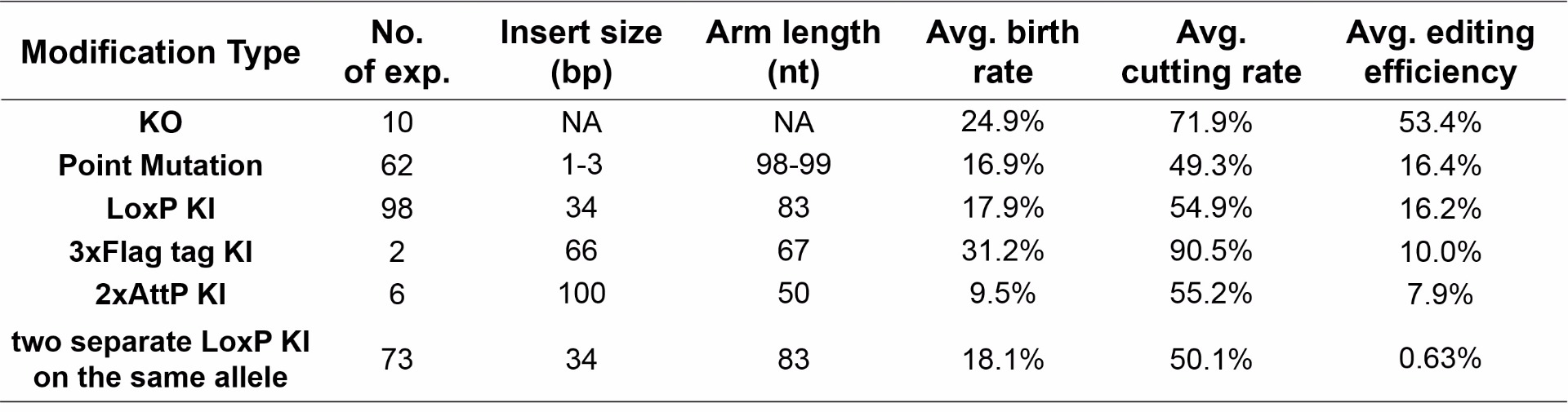
Gene editing efficiency and the birth rate is also affected by the size of the insert and length of the homologous regions when using single-stranded oligo donor DNA
A technical limitation of ssODN is its length. Most commercial vendors can manufacture up to 200 bases of single stranded DNA molecules. Therefore, homologous regions for a ssODN can be shortened if the insert size gets bigger. The system is perfect for point mutation insertions and can accommodate small fragment inserts up to 100 bp.
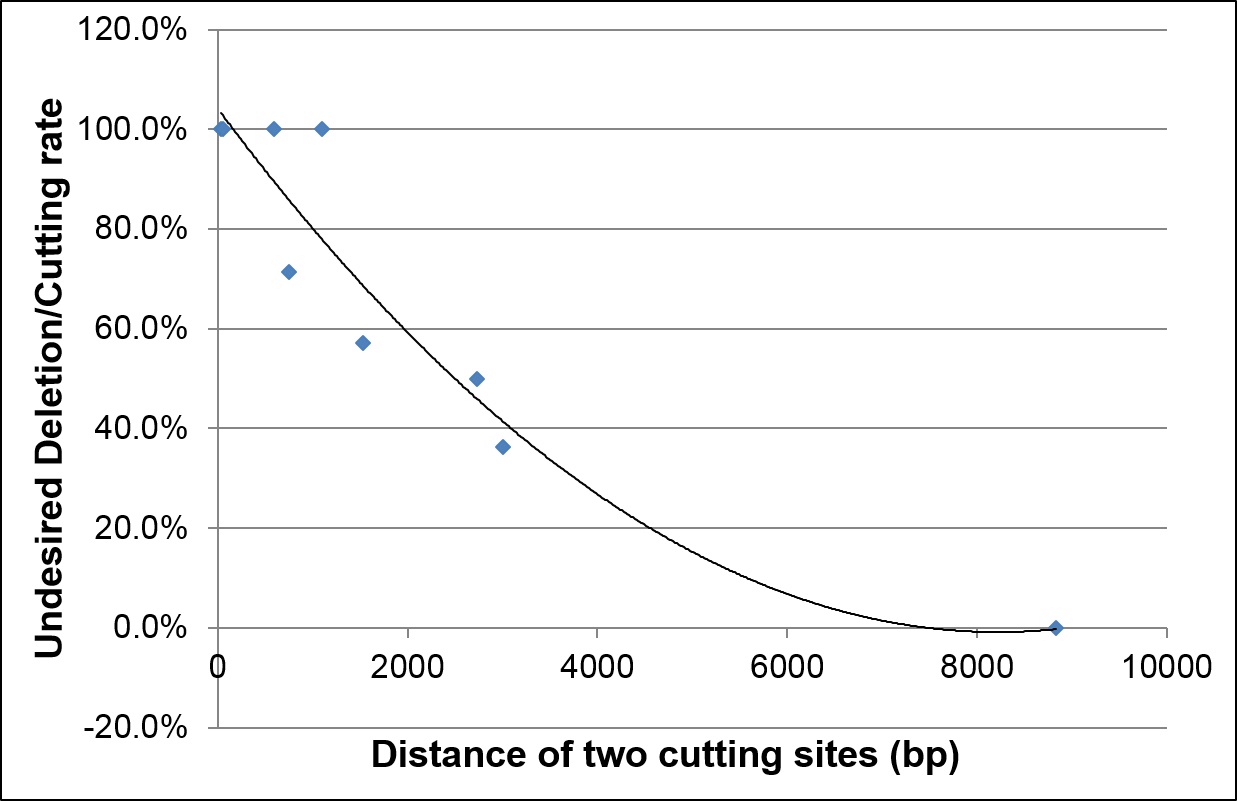
Distance between 2 cutting sites must be >3kb
In projects necessitating 2 modification sites such as generating a conditional knock-out model with 2 LoxP inserts, the distance between cutting sites should be at least 3 kb for simultaneous modification to avoid deletions between the two cut sites due to efficient NHEJ repair. If the distance between sites is <3 kb, sequential modification is recommended. The recommended distance between the two insertion sites should be >6 kb for efficient simultaneous modifications and to limit undesired deletion events.
Summary of our technical discussion about CRISPR gene modification in mouse models
- Optimal conditions are necessary for high efficiency generating mouse models
- Cas9/gRNA/donor DNA concentration; insert size and homology arm length are crucial for balancing cutting rate, gene editing efficiency, and birth rate
- The optimal distance between two cutting sites for simultaneous modification is >6 kb
Support Materials
Posters:
Publications
CRISPR Mouse/ Rat Models: Knock-in, Knockout, and Conditional Knockout
CRISPR Technology
-
Smalley, E. (2016). CRISPR mouse model boom, rat model renaissance. Nature Biotechnology. 34, 893–894.
-
Baker, M. (2014). Gene editing at CRISPR speed. Nature biotechnology, 32(4), 309-313.
CRISPR Knock-in H11 Locus in Pigs
-
Ruan, J., Li, H., Xu, K., Wu, T., Wei, J., Zhou, R., ... & Chen-Tsai, R. Y. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Scientific reports, 5, 14253.
Knock-in, Knockout, Conditional Knock-out
- Park, J., Jung, E., Lee, S. H., & Chung, W. S. (2020). CDC50A dependent phosphatidylserine exposure induces inhibitory post-synapse elimination by microglia. bioRxiv.
- Ramachandra Rao, S., Fliesler, S. J., Kotla, P., Nguyen, M. N., & Pittler, S. J. (2020). Lack of Overt Retinal Degeneration in a K42E Dhdds Knock-In Mouse Model of RP59. Cells, 9(4), 896.
- Beurg, M., Barlow, A., Furness, D. N., & Fettiplace, R. (2019). A Tmc1 mutation reduces calcium permeability and expression of mechanoelectrical transduction channels in cochlear hair cells. Proceedings of the National Academy of Sciences, 116(41), 20743-20749.
- Goldring, A. C., Beurg, M., & Fettiplace, R. (2019). The contribution of TMC1 to adaptation of mechanoelectrical transduction channels in cochlear outer hair cells. The Journal of physiology.
- Hwang, S., He, Y., Xiang, X., Seo, W., Kim, S. J., Ma, J., ... & Kunos, G. (2019). Interleukin‐22 ameliorates neutrophil‐driven nonalcoholic steatohepatitis through multiple targets. Hepatology https://doi.org/10.1002/hep.31031.
- Dumesic, P. A., Egan, D. F., Gut, P., Tran, M. T., Parisi, A., Chatterjee, N., ... & Dou, F. (2019). An Evolutionarily Conserved uORF Regulates PGC1α and Oxidative Metabolism in Mice, Flies, and Bluefin Tuna. Cell metabolism.
- Liang, T., Zhang, H., Xu, Q., Wang, S., Qin, C., & Lu, Y. (2019). Mutant Dentin Sialophosphoprotein Causes Dentinogenesis Imperfecta. Journal of dental research, 0022034519854029.
- Qian, W., Miner, C. A., Ingle, H., Platt, D. J., Baldridge, M. T., & Miner, J. J. (2019). A human STAT1 gain-of-function mutation impairs CD8+ T cell responses against gammaherpesvirus-68. Journal of virology, JVI-00307.
- Kweon, S. M., Chen, Y., Moon, E., Kvederaviciutė, K., Klimasauskas, S., & Feldman, D. E. (2019). An Adversarial DNA N6-Methyladenine-Sensor Network Preserves Polycomb Silencing. Molecular Cell. https://doi.org/10.1016/j.molcel.2019.03.018
- Deng, F., He, S., Cui, S., Shi, Y., Tan, Y., Li, Z., ... & Peng, L. (2018). A Molecular Targeted Immunotherapeutic Strategy for Ulcerative Colitis via Dual-Targeting Nanoparticles Delivering miR-146b to Intestinal Macrophages. Journal of Crohn's and Colitis.
- Jo, S., Fonseca, T. L., Bocco, B. M. D. C., Fernandes, G. W., McAninch, E. A., Bolin, A. P., ... & Németh, D. (2018). Type 2 deiodinase polymorphism causes ER stress and hypothyroidism in the brain. The Journal of Clinical Investigation.
- Langston, R. G., Rudenko, I. N., Kumaran, R., Hauser, D. N., Kaganovich, A., Ponce, L. B., ... & Beilina, A. (2018). Differences in Stability, Activity and Mutation Effects Between Human and Mouse Leucine-Rich Repeat Kinase 2. Neurochemical research, 1-14.
- Amara, N., Tholen, M., & Bogyo, M. (2018). Chemical tools for selective activity profiling of endogenously expressed MMP-14 in multicellular models. ACS Chemical Biology. doi: 10.1021/acschembio.8b00562.
- Allocca, S., Ciano, M., Ciardulli, M. C., D’Ambrosio, C., Scaloni, A., Sarnataro, D., ... & Bonatti, S. (2018). An αB-Crystallin Peptide Rescues Compartmentalization and Trafficking Response to Cu Overload of ATP7B-H1069Q, the Most Frequent Cause of Wilson Disease in the Caucasian Population. International journal of molecular sciences, 19(7).
-
*Peng, L., Zhang, H., Hao, Y., Xu, F., Yang, J., Zhang, R., ... & Chen, C. (2016). Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine, 14, 83-96.
-
*Hu, J. K., Crampton, J. C., Locci, M., & Crotty, S. (2016). CRISPR-mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ triple gene disruption reveals NKT cell defects but not T follicular helper cell defects. PloS one, 11(5), e0156074.
-
*Besschetnova, T. Y., Ichimura, T., Katebi, N., Croix, B. S., Bonventre, J. V., & Olsen, B. R. (2015). Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biology, 42, 56-73.
-
*McKenzie, C. W., Craige, B., Kroeger, T. V., Finn, R., Wyatt, T. A., Sisson, J. H., ... & Lee, L. (2015). CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Molecular biology of the cell, 26(18), 3140-3149.
-
*Bishop, K. A., Harrington, A., Kouranova, E., Weinstein, E. J., Rosen, C. J., Cui, X., & Liaw, L. (2016). CRISPR/Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3: Genes, Genomes, Genetics, 6(7), 2051-2061.
For more journal references, please visit our comprehensive list of citations and reference publications.
FAQs
How many embryos are microinjected for validation of gRNAs?
What is the correlation between cutting in the embryo test (in vivo gRNA validation) and the final mouse?
Which technology is suitable for inserting a gene of interest with GFP or Cre-ERT?




