Newsletter
TARGATT™ Master Cell Line Generation Service
Applied StemCell’s proprietary site-specific, integrase-based TARGATT™ technology has been used to generate stable, high-quality TARGATT™ Master iPS, CHO, and HEK293 cell lines for large transgenes knock-in. However, if you would like to knock in your gene-of-interest (GOI) into your own cell line, our experts can engineer the TARGATT™ landing pad into your cells. With your engineered TARGATT™ cell line and our unique integrase, you can insert a single copy of your DNA fragment (up to 20kb) at a safe harbor locus. If you would like to learn more, contact us today to schedule your free consultation.
- Fully customizable service
- Our experts can help enhance your project design
- Generate your TARGATT™"Master" cells for reporter/tag line development, conditional gene expression models, and more
- Free consultation
- Fast turn around
- Off-the-shelf TARGATT™ Master iPS, CHO, and HEK293 Cell Lines are available
- TARGATT™ Advantages
- Single copy knockin: 1 cell, 1 docking site, 1 inserted transgene
- Site-specific knockin into a high expression, safe harbor locus (H11, ASC2)
- High-efficiency integration
- Large cell library construction in HEK293/CHO cells
- Cost-effective: no virus packaging time and resources
Products and Services
Technical Details

Applied StemCell’s proprietary TARGATT™ technology allows for the site-specific integration of large DNA fragments. The TARGATT™ technology uses a serine integrase to mediate an irreversible recombination between a pre-engineered “attP” docking-site and an attP recognition-sequence on the donor vector “attB”, for stable gene integration. The attP docking sites can be engineered into a preselected locus, such as an intergenic, transcriptionally active safe harbor locus, using CRISPR/Cas9. The irreversible transgene integration mediated by the integrase ensures a high efficiency and stability of integration.
Applications:
- Precise comparison of different genes from more than one cell line
- Generate a master cell line expressing different reporter genes
- Gene overexpression
- Inducible gene expression models
Applied StemCell is now providing a TARGATT™ master knock-in cell line generation service
-
You send us your favorite cell lines
-
We will insert the attP docking sites at specific loci
-
Fast: get your transgenic cell line in 3 months!
Application Notes
Combining CRISPR & TARGATT™ in hiPSCs to Enable Large DNA Insertion
1. CRISPR: Insertion of 70 bp attP docking site to create TARGATT™ Master Cell Line
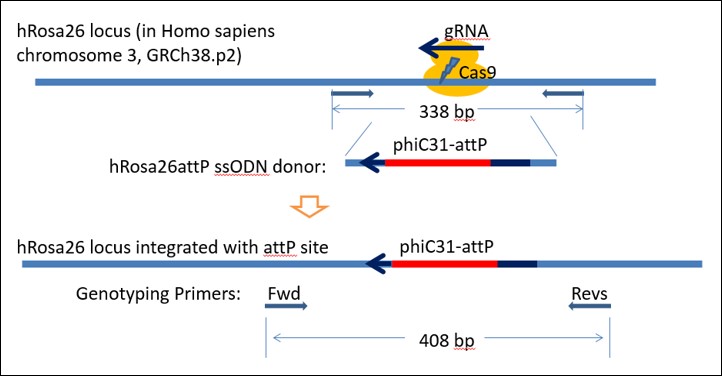
Figure 1. Strategy to knock-in attP "docking sites" into hiPSC Rosa26 locus to generate a TARGATT™ iPSC Master Cell Line
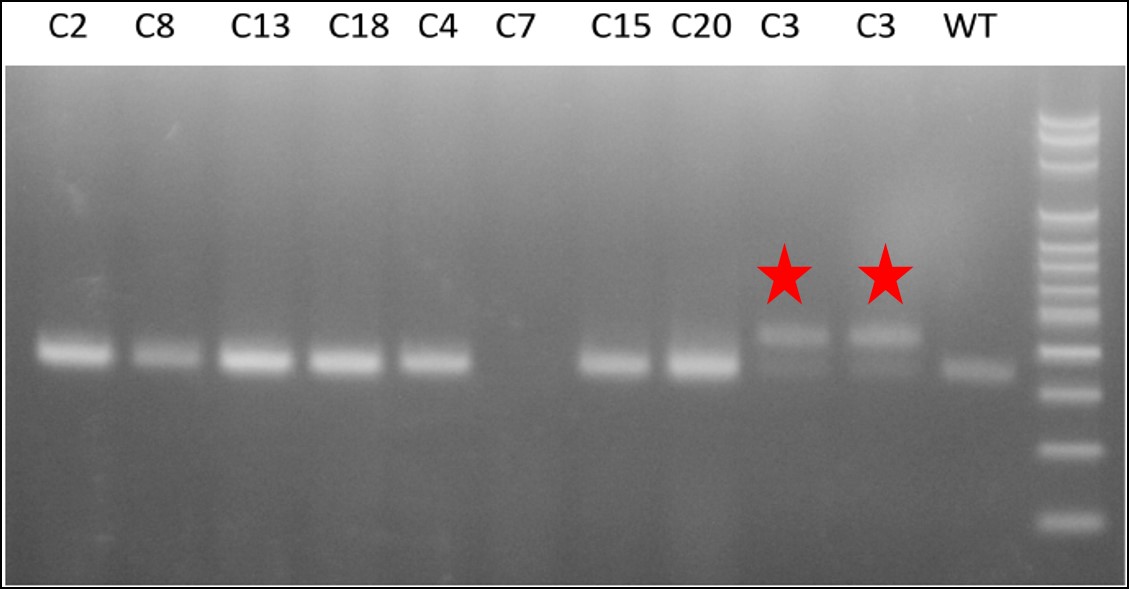
Figure 2. Clone 3 shows heterozygous insertion of attP sequence.
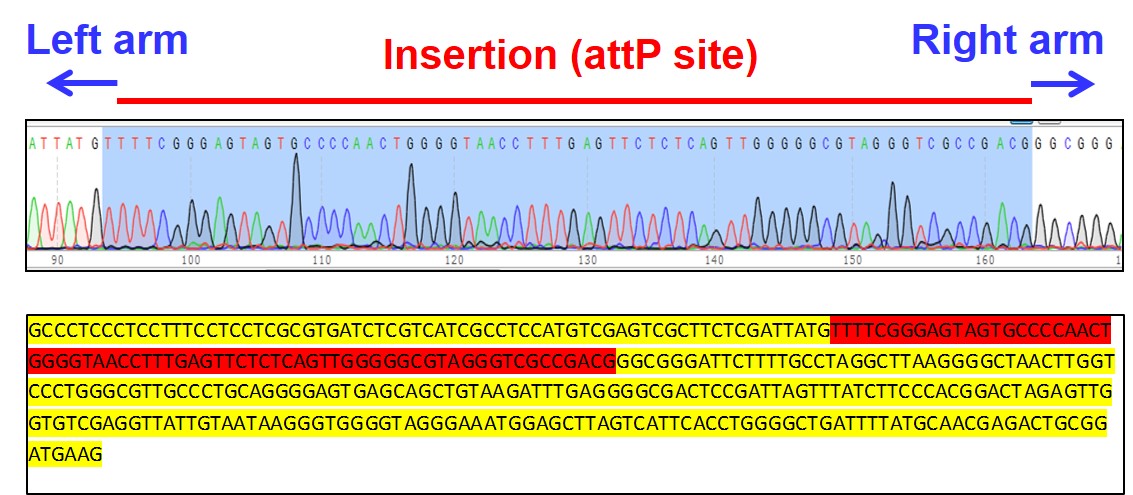
Figure 3. Sequencing of clone C3 shows insertion of 70 bp attP docking site in Rosa26 locus (red)
2. TARGATT™: Large DNA Insertion in iPSC Master Cell Line
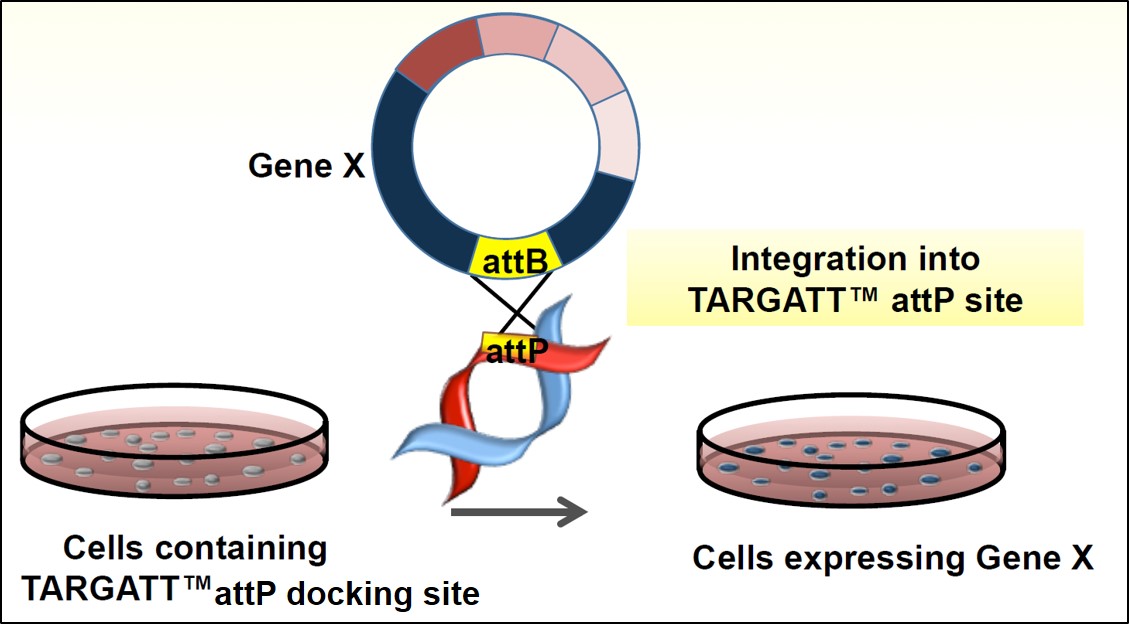
Figure 4. Schematic illustration of the integrase based knock-in in hiPS cells
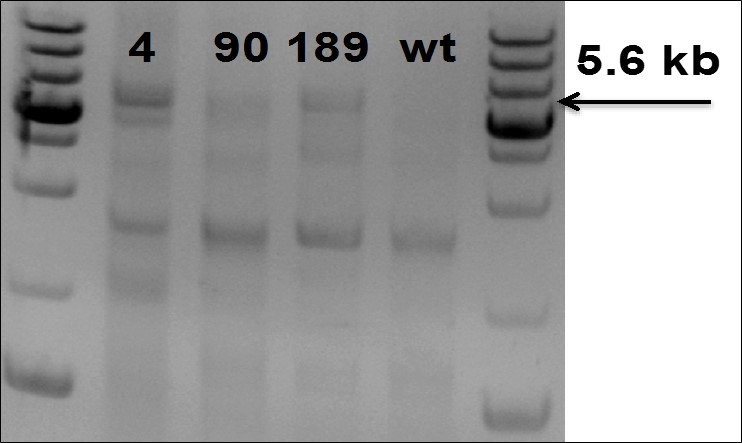
Figure 5. PCR Gel electrophoresis to confirm insertion of 5.6 kb fragment in TARGATT™ iPS Master cell line.
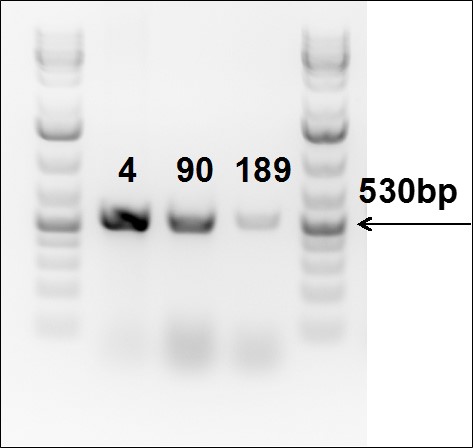
Figure 6. PCR to confirm gene knock-in into Rosa26 locus in TARGATT™ Master iPSC line.
Site-Specific Knock-in of TARGATT™ sites
.jpg)
Figure: Using mCherry reporter (left) in human iPSC line. Original Cell Line: hiPSC (right).
Support Materials
Brochures/ Flyers:
Posters:
Webinar:
![]() TARGATT™: A Mammalian System for Antibody/ Protein Library Screening (October 2018)
TARGATT™: A Mammalian System for Antibody/ Protein Library Screening (October 2018)![]() TARGATT™ and CRISPR/Cas9 modified induced pluripotent stem cells (iPSCs) for in vitro genetic disease modeling
TARGATT™ and CRISPR/Cas9 modified induced pluripotent stem cells (iPSCs) for in vitro genetic disease modeling
Publications
Master Cell Line
- Chi, X., Zheng, Q., Jiang, R., Chen-Tsai, R. Y., & Kong, L. J. (2019). A system for site-specific integration of transgenes in mammalian cells. PLOS ONE, 14(7), e0219842.
Mouse Book Chapters
- Chen-Tsai, R. Y. (2020). Integrase-Mediated Targeted Transgenics Through Pronuclear Microinjection. In Transgenic Mouse (pp. 35-46). Humana, New York, NY.
- Chen-Tsai, R. Y. (2019). Using TARGATT™ Technology to Generate Site-Specific Transgenic Mice. In Microinjection (pp. 71-86). Humana Press, New York, NY.
Description of the technology
- Zhu, F., Gamboa, M., Farruggio, A. P., Hippenmeyer, S., Tasic, B., Schüle, B., … Calos, M. P. (2014). DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Research, 42(5), e34. http://doi.org/10.1093/nar/gkt1290.
- Tasic, B., Hippenmeyer, S., Wang, C., Gamboa, M., Zong, H., Chen-Tsai, Y., & Luo, L. (2011). Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proceedings of the National Academy of Sciences of the United States of America, 108(19), 7902–7907. http://doi.org/10.1073/pnas.1019507108.
Commentary, comparison with other transgenic methods
- Rossant, J., Nutter, L. M., & Gertsenstein, M. (2011). Engineering the embryo. Proceedings of the National Academy of Sciences, 108(19), 7659-7660.
Tet inducible mice generated by TARGATT™
- Fan, X., Petitt, M., Gamboa, M., Huang, M., Dhal, S., Druzin, M. L., … Nayak, N. R. (2012). Transient, Inducible, Placenta-Specific Gene Expression in Mice. Endocrinology, 153(11), 5637–5644. http://doi.org/10.1210/en.2012-1556.
Advantage of Hipp11 (H11) locus
- Hippenmeyer, S., Youn, Y. H., Moon, H. M., Miyamichi, K., Zong, H., Wynshaw-Boris, A., & Luo, L. (2010). Genetic Mosaic Dissection of Lis1 and Ndel1 in Neuronal Migration. Neuron, 68(4), 695–709. http://doi.org/10.1016/j.neuron.2010.09.027.
Applications for mice generated by TARGATT™ (and cited/published articles)
- Lindtner, S., Catta-Preta, R., Tian, H., Su-Feher, L., Price, J. D., Dickel, D. E., ... & Pennacchio, L. A. (2019). Genomic Resolution of DLX-Orchestrated Transcriptional Circuits Driving Development of Forebrain GABAergic Neurons. Cell reports, 28(8), 2048-2063.
- Wang, T. A., Teo, C. F., Åkerblom, M., Chen, C., Tynan-La Fontaine, M., Greiner, V. J., ... & Jan, L. Y. (2019). Thermoregulation via Temperature-Dependent PGD2 Production in Mouse Preoptic Area. Neuron, 103(2), 309-322.
- Clarke, B. A., Majumder, S., Zhu, H., Lee, Y. T., Kono, M., Li, C., ... & Byrnes, C. (2019). The Ormdl genes regulate the sphingolipid synthesis pathway to ensure proper myelination and neurologic function in mice. eLife, 8.
- Carlson, H. L., & Stadler, H. S. (2019). Development and functional characterization of a lncRNA‐HIT conditional loss of function allele. genesis, e23351.
- Chande, S., Ho, B., Fetene, J., & Bergwitz, C. (2019). Transgenic mouse model for conditional expression of influenza hemagglutinin-tagged human SLC20A1/PIT1. PloS one, 14(10), e0223052. doi:10.1371/journal.pone.0223052
- Hu, Q., Ye, Y., Chan, L. C., Li, Y., Liang, K., Lin, A., ... & Pan, Y. (2019). Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nature immunology, 1.
- Matharu, N., Rattanasopha, S., Tamura, S., Maliskova, L., Wang, Y., Bernard, A., ... & Ahituv, N. (2018). CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science, eaau0629.
- Barrett, R. D., Laurent, S., Mallarino, R., Pfeifer, S. P., Xu, C. C., Foll, M., ... & Hoekstra, H. E. (2018). The fitness consequences of genetic variation in wild populations of mice. bioRxiv, 383240.
- Ibrahim, L. A., Huang, J. J., Wang, S. Z., Kim, Y. J., Li, I., & Huizhong, W. (2018). Sparse Labeling and Neural Tracing in Brain Circuits by STARS Strategy: Revealing Morphological Development of Type II Spiral Ganglion Neurons. Cerebral Cortex, 1-14.
- Kumar, A., Dhar, S., Campanelli, G., Butt, N. A., Schallheim, J. M., Gomez, C. R., & Levenson, A. S. (2018). MTA 1 drives malignant progression and bone metastasis in prostate cancer. Molecular oncology.
- Jang, Y., Wang, C., Broun, A., Park, Y. K., Zhuang, L., Lee, J. E., ... & Ge, K. (2018). H3. 3K4M destabilizes enhancer epigenomic writers MLL3/4 and impairs adipose tissue development. bioRxiv, 301986. doi:https://doi.org/10.1101/301986
- Tang, Y., Kwon, H., Neel, B. A., Kasher-Meron, M., Pessin, J., Yamada, E., & Pessin, J. E. (2018). The fructose-2, 6-bisphosphatase TIGAR suppresses NF-κB signaling by directly inhibiting the linear ubiquitin assembly complex LUBAC. Journal of Biological Chemistry, jbc-RA118.
- Chen, M., Geoffroy, C. G., Meves, J. M., Narang, A., Li, Y., Nguyen, M. T., ... & Elzière, L. (2018). Leucine Zipper-Bearing Kinase Is a Critical Regulator of Astrocyte Reactivity in the Adult Mammalian CNS. Cell Reports, 22(13), 3587-3597.
- Kido, T., Sun, Z., & Lau, Y.-F. C. (2017). Aberrant activation of the human sex-determining gene in early embryonic development results in postnatal growth retardation and lethality in mice. Scientific Reports, 7, 4113. http://doi.org/10.1038/s41598-017-04117-6.
- Nouri, N., & Awatramani, R. (2017). A novel floor plate boundary defined by adjacent En1 and Dbx1 microdomains distinguishes midbrain dopamine and hypothalamic neurons. Development, 144(5), 916-927.
- Li, K., Wang, F., Cao, W. B., Lv, X. X., Hua, F., Cui, B., ... & Yu, J. M. (2017). TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of p53-Mediated Senescence. Cancer Cell, 31(5), 697-710.
- Matharu, N., Rattanasopha, S., Maliskova, L., Wang, Y., Hardin, A., Vaisse, C., & Ahituv, N. (2017). Promoter or Enhancer Activation by CRISPRa Rescues Haploinsufficiency Caused Obesity. bioRxiv, 140426.
- Jiang, T., Kindt, K., & Wu, D. K. (2017). Transcription factor Emx2 controls stereociliary bundle orientation of sensory hair cells. eLife, 6, e23661.
- Booze, M. L., Hansen, J. M., & Vitiello, P. F. (2016). A Novel Mouse Model for the Identification of Thioredoxin-1 Protein Interactions. Free Radical Biology & Medicine, 99, 533–543. http://doi.org/10.1016/j.freeradbiomed.2016.09.013.
- Feng, D., Dai, S., Liu, F., Ohtake, Y., Zhou, Z., Wang, H., ... & Hayat, U. (2016). Cre-inducible human CD59 mediates rapid cell ablation after intermedilysin administration. The Journal of clinical investigation, 126(6), 2321-2333.
- Sun, N., Yun, J., Liu, J., Malide, D., Liu, C., Rovira, I. I., … Finkel, T. (2015). Measuring in vivo mitophagy. Molecular Cell, 60(4), 685–696. http://doi.org/10.1016/j.molcel.2015.10.009.
- Devine, W. P., Wythe, J. D., George, M., Koshiba-Takeuchi, K., & Bruneau, B. G. (2014). Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife, 3, e03848. http://doi.org/10.7554/eLife.03848.
- Fogg, P. C. M., Colloms, S., Rosser, S., Stark, M., & Smith, M. C. M. (2014). New Applications for Phage Integrases. Journal of Molecular Biology, 426(15), 2703–2716. http://doi.org/10.1016/j.jmb.2014.05.014.
- Chen-Tsai, R. Y., Jiang, R., Zhuang, L., Wu, J., Li, L., & Wu, J. (2014). Genome editing and animal models. Chinese science bulletin, 59(1), 1-6.
- Park, K.-E., Park, C.-H., Powell, A., Martin, J., Donovan, D. M., & Telugu, B. P. (2016). Targeted Gene Knockin in Porcine Somatic Cells Using CRISPR/Cas Ribonucleoproteins. International Journal of Molecular Sciences, 17(6), 810. http://doi.org/10.3390/ijms17060810.
- Guenther, C. A., Tasic, B., Luo, L., Bedell, M. A., & Kingsley, D. M. (2014). A molecular basis for classic blond hair color in Europeans. Nature Genetics, 46(7), 748–752. http://doi.org/10.1038/ng.2991.
- Villamizar, C. A. (2014). Characterization of the vascular pathology in the acta2 r258c mouse model and cerebrovascular characterization of the acta2 null mouse. UT GSBS Dissertations and These (Open Access). Paper 508 (2014)
FAQs
I am interested in making a master TARGATT™ cell line using my own cell line as a parental cell line. Can you take my cancer cell line, for example, to generate a master cell line using your service?
What is the Gene Knock-in Technology used to generate master cell lines?
How long does it take to make a master cell line?
Is there a size limit on DNA to be inserted into the genome (to attP site)?
What is the final deliverable product?
What safe harbor loci are available to place the TARGATT™ docking site? Can you recommend one for my cell lines?
I got my master cell line. Do you provide plasmid so that we can construct Knock-in vector with our gene of interest?
Do you have off-the-shelf TARGATT™ Master cell lines?