Newsletter
Bioproduction: CHO Cells
TARGATT™ CHO Cells for Recombinant Protein and Antibody Bioproduction! Applied StemCell’s TARGATT™ Master CHO cell lines leverage the site-specific gene integration capacity of serine integrase, phiC31 to knockin transgene(s) into transcriptionally active genomic hotspots, such as the H11 or the proprietary ASC2 (A2) safe harbor locus. These master cell lines enable highly efficient and rapid gene insertion and high levels of recombinant protein/antibody expression.
Our Master TARGATT™ CHO cell lines can exceed the capabilities of the traditionally made CHO antibody production cells and offers a platform for affordable and feasible bioproduction for antibodies and other recombinant proteins for companies and projects of all sizes.
- High knock-in efficiency
- Site-specific integration into a well-defined safe harbor locus
- Single gene knocki-n: one variant - one locus - one cell line
- Unidirectional integration for stable knock-in cell lines
- Uniform, high-level gene expression
- Overcomes challenges such as random insertion, gene silencing, multiple copy gene integration, ablated gene expression.
Licensing and Evaluation Programs Available!
Products and Services
Technical Details
ASC’s TARGATT™ technology improves CHO recombinant protein or antibody production capability by reducing cost and improving manufacturing feasibility for companies and projects of all sizes. Please let us know if you have questions about your CHO bioproduction project.
|
Features |
TARGATT™ Site-Specific Insertion |
Traditional Random Gene Insertion |
|
Safe Genomic Locus |
Yes |
No; random |
|
Protein Yield |
High |
Varies |
|
Gene Insertion Efficiency |
High |
Low |
|
Reproducibility |
Yes; for any protein |
No; different from one protein to another |
|
Copy # of Inserted Gene |
Single copy |
Multiple copies |
|
Stable |
Yes |
No; Chromosome rearrangement |
|
Gene Silencing |
No |
Yes |
Advantages of using TARGATT™ Master Cell Lines for gene knockin:
- Stable cell line generation
- Single copy, site-specific gene integration into the safe harbor locus
- Guaranteed gene expression from an active, intergenic locus
- Footprint-free integration of gene of interest
- Use of selection marker is not necessary but optional
- Easy-to-use protocol requiring transfection of only one (donor) plasmid
Key points:
- Ability to achieve > 1 g/l antibody bioproduction using a single site insertion with little optimization
- Identification of the ASC-2 locus has allowed for a further > 2.5-fold increase in protein expression levels
TARGATT™ System in CHO Cells
Uniform Expression of Transgene in TARGATT™ CHO Cells
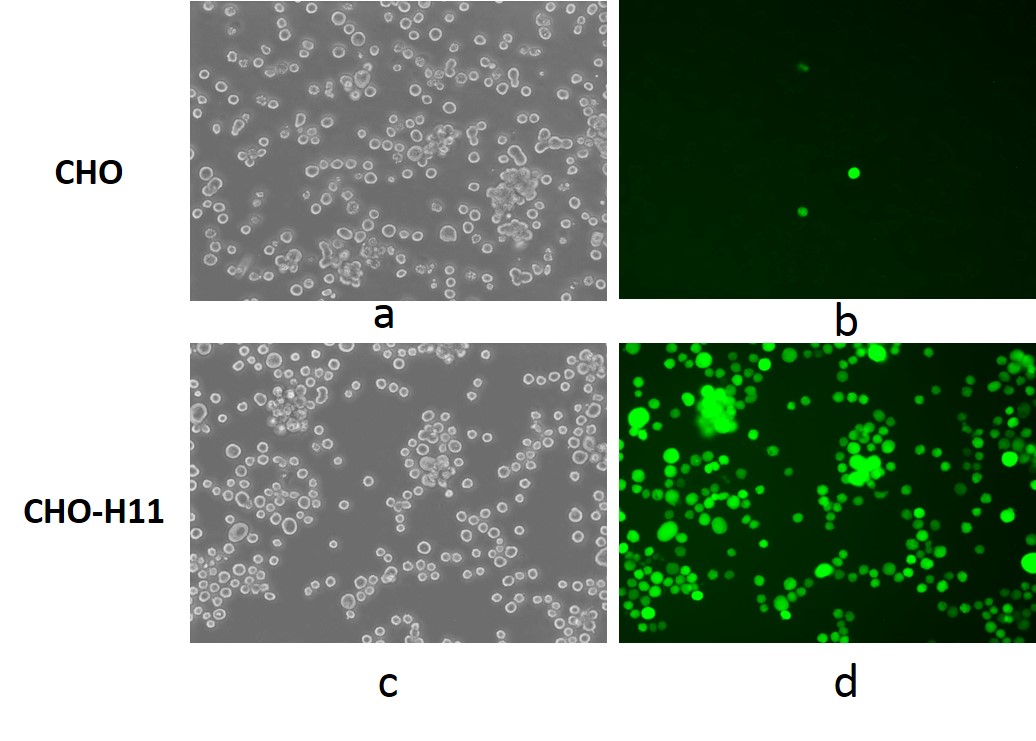
Figure 1. TARGATT™ CHO Master Cell Line with GFP transgene integration into the H11 locus. GFP signal was detected by fluorescence imaging after transfection with donor construct by random insertion in bright field (a) and GFP channel (b), and TARGATT™ integration plus GCV selection in bright field (c) and GFP channel (d).
High Efficiency GFP Knockin and Expression in TARGATT™ CHO Cells
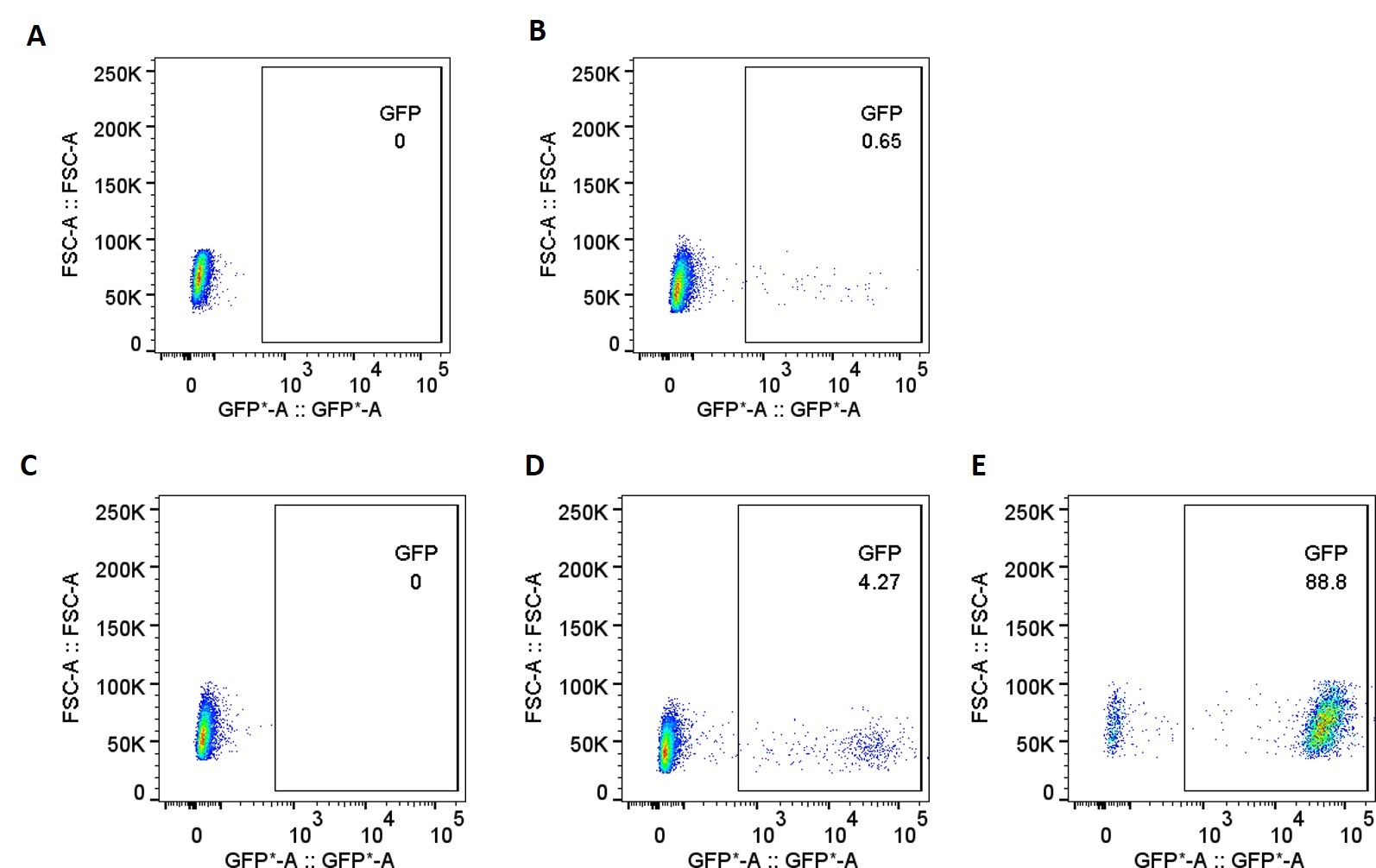
Figure 2. GFP expression was measured by fluorescence imaging after transfection in parental CHO-K1 cell lines by random integration (A-B) and in TARGATT™ CHO-K1 Master Cell Line by site-specific gene integration (C-E). (A) CHO-K1 parental cell line without transfection; (B) CHO-K1 parental cell line randomly transfected with GFP plasmid; (C) CHO-K1 master cell line without transfection; (D) CHO-K1 master cell line transfected with GFP plasmid before GCV selection; (E) CHO-K1 master cell line transfected with GFP plasmid after GCV selection.
Bioproduction in TARGATT™ CHO Cells
Antibody Production in TARGATT™ CHO (H11) Cells
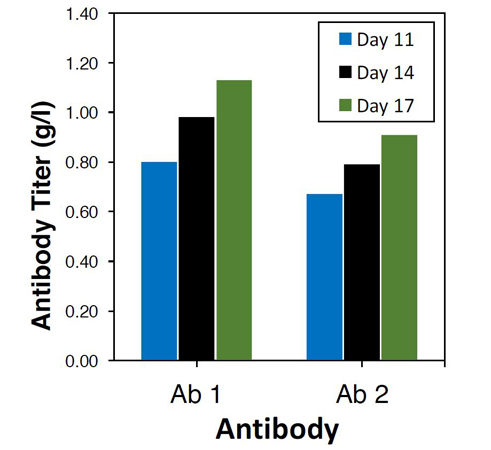
Figure 1. Expression of antibodies Ab1 and Ab2 in the H11 genomic hotspot locus of the TARGATT™ CHO (H11) cells yielded ~1g/L of each protein.
Optimizing Docking Site Location in TARGATT™ CHO Cell Lines
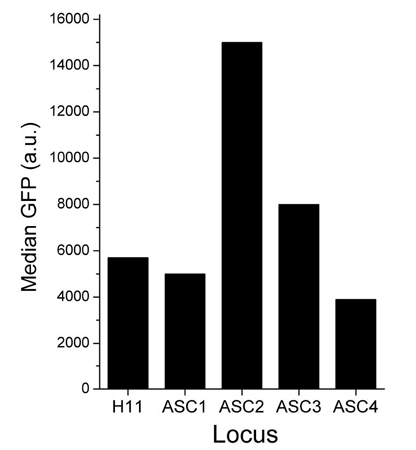
Figure 2. Expression of green fluorescent protein (GFP) in various genomic hotspot loci in TARGATT™ CHO cells. TARGATT™ CHO master cell lines were engineered with an attP docking site in the well-characterized H11 locus and 4 different proprietary loci, ASC1, ASC2, ASC3, and ASC4. The GFP reporter gene was inserted into each cell line and its expression from each locus was measured using FACS analysis. Reporter expression in the ASC2 (A2) locus was 2.5-fold higher than the expression in the H11 locus. The estimated protein yield from the TARGATT™ CHO (A2) Master cell line is ~2.5 g/L.
eBook: TARGATT™ Technology For Antibody Discovery and Screening (March 2021)
*Featured in Informa Connect's eBook: Antibody Discovery, Selection & Screening.
Support Materials
eBook:
TARGATT™ Technology For Antibody Discovery and Screening
(March 2021)
*Featured in Informa Connect's eBook: Antibody
Discovery, Selection & Screening.
Poster:
![]() TARGATT™: Rapid and Efficient Transgene Integration Technology to Develop Large Mammalian Cell-Based Screening Libraries
TARGATT™: Rapid and Efficient Transgene Integration Technology to Develop Large Mammalian Cell-Based Screening Libraries
Webinar:
![]() TARGATT™ Rapid and Efficient Transgene Integration Technology: Stable Cell Line Development and Generation of Large Mammalian Cell-based Screening Libraries
TARGATT™ Rapid and Efficient Transgene Integration Technology: Stable Cell Line Development and Generation of Large Mammalian Cell-based Screening Libraries
Publications
TARGATT™ Master Cell Line
- Chi, X., Zheng, Q., Jiang, R., Chen-Tsai, R. Y., & Kong, L. J. (2019). A system for site-specific integration of transgenes in mammalian cells. PLOS ONE, 14(7), e0219842.
Transgenic Mouse Book Chapters
- Chen-Tsai, R. Y. (2020). Integrase-Mediated Targeted Transgenics Through Pronuclear Microinjection. In Transgenic Mouse (pp. 35-46). Humana, New York, NY.
- Chen-Tsai, R. Y. (2019). Using TARGATT™ Technology to Generate Site-Specific Transgenic Mice. In Microinjection (pp. 71-86). Humana Press, New York, NY.
Description of the technology
- Zhu, F., Gamboa, M., Farruggio, A. P., Hippenmeyer, S., Tasic, B., Schüle, B., … Calos, M. P. (2014). DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Research, 42(5), e34. http://doi.org/10.1093/nar/gkt1290.
- Tasic, B., Hippenmeyer, S., Wang, C., Gamboa, M., Zong, H., Chen-Tsai, Y., & Luo, L. (2011). Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proceedings of the National Academy of Sciences of the United States of America, 108(19), 7902–7907. http://doi.org/10.1073/pnas.1019507108.
Commentary, comparison with other transgenic methods
- Rossant, J., Nutter, L. M., & Gertsenstein, M. (2011). Engineering the embryo. Proceedings of the National Academy of Sciences, 108(19), 7659-7660.
Tet inducible mice generated by TARGATT™
- Fan, X., Petitt, M., Gamboa, M., Huang, M., Dhal, S., Druzin, M. L., … Nayak, N. R. (2012). Transient, Inducible, Placenta-Specific Gene Expression in Mice. Endocrinology, 153(11), 5637–5644. http://doi.org/10.1210/en.2012-1556.
Advantage of Hipp11 (H11) locus
- Hippenmeyer, S., Youn, Y. H., Moon, H. M., Miyamichi, K., Zong, H., Wynshaw-Boris, A., & Luo, L. (2010). Genetic Mosaic Dissection of Lis1 and Ndel1 in Neuronal Migration. Neuron, 68(4), 695–709. http://doi.org/10.1016/j.neuron.2010.09.027.
Applications for TARGATT™ technology
- Lindtner, S., Catta-Preta, R., Tian, H., Su-Feher, L., Price, J. D., Dickel, D. E., ... & Pennacchio, L. A. (2019). Genomic Resolution of DLX-Orchestrated Transcriptional Circuits Driving Development of Forebrain GABAergic Neurons. Cell reports, 28(8), 2048-2063.
- Wang, T. A., Teo, C. F., Åkerblom, M., Chen, C., Tynan-La Fontaine, M., Greiner, V. J., ... & Jan, L. Y. (2019). Thermoregulation via Temperature-Dependent PGD2 Production in Mouse Preoptic Area. Neuron, 103(2), 309-322.
- Clarke, B. A., Majumder, S., Zhu, H., Lee, Y. T., Kono, M., Li, C., ... & Byrnes, C. (2019). The Ormdl genes regulate the sphingolipid synthesis pathway to ensure proper myelination and neurologic function in mice. eLife, 8.
- Carlson, H. L., & Stadler, H. S. (2019). Development and functional characterization of a lncRNA‐HIT conditional loss of function allele. genesis, e23351.
- Chande, S., Ho, B., Fetene, J., & Bergwitz, C. (2019). Transgenic mouse model for conditional expression of influenza hemagglutinin-tagged human SLC20A1/PIT1. PloS one, 14(10), e0223052. doi:10.1371/journal.pone.0223052
- Hu, Q., Ye, Y., Chan, L. C., Li, Y., Liang, K., Lin, A., ... & Pan, Y. (2019). Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nature immunology, 1.
- Matharu, N., Rattanasopha, S., Tamura, S., Maliskova, L., Wang, Y., Bernard, A., ... & Ahituv, N. (2018). CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science, eaau0629.
- Chen-Tsai, R. Y. (2019). Using TARGATT™ Technology to Generate Site-Specific Transgenic Mice. In Microinjection (pp. 71-86). Humana Press, New York, NY
- Barrett, R. D., Laurent, S., Mallarino, R., Pfeifer, S. P., Xu, C. C., Foll, M., ... & Hoekstra, H. E. (2018). The fitness consequences of genetic variation in wild populations of mice. bioRxiv, 383240.
- Ibrahim, L. A., Huang, J. J., Wang, S. Z., Kim, Y. J., Li, I., & Huizhong, W. (2018). Sparse Labeling and Neural Tracing in Brain Circuits by STARS Strategy: Revealing Morphological Development of Type II Spiral Ganglion Neurons. Cerebral Cortex, 1-14.
- Kumar, A., Dhar, S., Campanelli, G., Butt, N. A., Schallheim, J. M., Gomez, C. R., & Levenson, A. S. (2018). MTA 1 drives malignant progression and bone metastasis in prostate cancer. Molecular oncology.
- Jang, Y., Broun, A., Wang, C., Park, Y. K., Zhuang, L., Lee, J. E., ... & Ge, K. (2018). H3. 3K4M destabilizes enhancer H3K4 methyltransferases MLL3/MLL4 and impairs adipose tissue development. Nucleic acids research. https://doi.org/10.1093/nar/gky982
- Tang, Y., Kwon, H., Neel, B. A., Kasher-Meron, M., Pessin, J., Yamada, E., & Pessin, J. E. (2018). The fructose-2, 6-bisphosphatase TIGAR suppresses NF-κB signaling by directly inhibiting the linear ubiquitin assembly complex LUBAC. Journal of Biological Chemistry, jbc-RA118.
- Chen, M., Geoffroy, C. G., Meves, J. M., Narang, A., Li, Y., Nguyen, M. T., ... & Elzière, L. (2018). Leucine Zipper-Bearing Kinase Is a Critical Regulator of Astrocyte Reactivity in the Adult Mammalian CNS. Cell Reports, 22(13), 3587-3597
- Kido, T., Sun, Z., & Lau, Y.-F. C. (2017). Aberrant activation of the human sex-determining gene in early embryonic development results in postnatal growth retardation and lethality in mice. Scientific Reports, 7, 4113. http://doi.org/10.1038/s41598-017-04117-6.
- Nouri, N., & Awatramani, R. (2017). A novel floor plate boundary defined by adjacent En1 and Dbx1 microdomains distinguishes midbrain dopamine and hypothalamic neurons. Development, 144(5), 916-927.
- Li, K., Wang, F., Cao, W. B., Lv, X. X., Hua, F., Cui, B., ... & Yu, J. M. (2017). TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of p53-Mediated Senescence. Cancer Cell, 31(5), 697-710.
- Jiang, T., Kindt, K., & Wu, D. K. (2017). Transcription factor Emx2 controls stereociliary bundle orientation of sensory hair cells. eLife, 6, e23661.
- Booze, M. L., Hansen, J. M., & Vitiello, P. F. (2016). A Novel Mouse Model for the Identification of Thioredoxin-1 Protein Interactions. Free Radical Biology & Medicine, 99, 533–543. http://doi.org/10.1016/j.freeradbiomed.2016.09.013.
- Feng, D., Dai, S., Liu, F., Ohtake, Y., Zhou, Z., Wang, H., ... & Hayat, U. (2016). Cre-inducible human CD59 mediates rapid cell ablation after intermedilysin administration. The Journal of clinical investigation, 126(6), 2321-2333.
- Sun, N., Yun, J., Liu, J., Malide, D., Liu, C., Rovira, I. I., … Finkel, T. (2015). Measuring in vivo mitophagy. Molecular Cell, 60(4), 685–696. http://doi.org/10.1016/j.molcel.2015.10.009.
- Devine, W. P., Wythe, J. D., George, M., Koshiba-Takeuchi, K., & Bruneau, B. G. (2014). Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife, 3, e03848. http://doi.org/10.7554/eLife.03848.
- Fogg, P. C. M., Colloms, S., Rosser, S., Stark, M., & Smith, M. C. M. (2014). New Applications for Phage Integrases. Journal of Molecular Biology, 426(15), 2703–2716. http://doi.org/10.1016/j.jmb.2014.05.014.
- Chen-Tsai, R. Y., Jiang, R., Zhuang, L., Wu, J., Li, L., & Wu, J. (2014). Genome editing and animal models. Chinese science bulletin, 59(1), 1-6.
- Park, K.-E., Park, C.-H., Powell, A., Martin, J., Donovan, D. M., & Telugu, B. P. (2016). Targeted Gene Knockin in Porcine Somatic Cells Using CRISPR/Cas Ribonucleoproteins. International Journal of Molecular Sciences, 17(6), 810. http://doi.org/10.3390/ijms17060810.
- Guenther, C. A., Tasic, B., Luo, L., Bedell, M. A., & Kingsley, D. M. (2014). A molecular basis for classic blond hair color in Europeans. Nature Genetics, 46(7), 748–752. http://doi.org/10.1038/ng.2991.
- Villamizar, C. A. (2014). Characterization of the vascular pathology in the acta2 r258c mouse model and cerebrovascular characterization of the acta2 null mouse. UT GSBS Dissertations and These (Open Access). Paper 508 (2014)
FAQs
What is the maximum size of DNA fragment for integration?
What are the differences between all your CHO knock-in kits?
Would multiple copies of transgene integration with random integration express more amount of protein than the TARGATT™ technology with only one copy of integrated transgene?
How does the level of expression with TARGATT™ CHO Master Cell Lines (the mentioned, ~1g/L、~2.5 g/L of protein) compare to random integration and other technologies?
What is the turnaround time for maximal expression compared with other technologies?
Is there any concern about leaky expression in the TARGATT™ CHO-K1 Master Cell Lines as seen in other technologies/ systems due to the presence of tetracycline in fetal bovine serum (FBS)?
Will you share the detailed sequence map of the donor plasmid with annotated sequences between the two attB sites (attB-Promoter-GOI-polyA-attB) for research where the promoter/ insulator needs to be replaced?
I want to do some culture of the CHO cells I got from you at a single cell level which means sorting cells into a 96 well plates, one cell per well, and manage to grow cells out of this. Do you culture cells on a single cell basis? Culture conditions?
Do your CHO cells (AST-1400, 1405, AST-1200) grow in adherent conditions?