Newsletter
ASC is a leading provider of genetically engineered mouse models for biomedical research and preclinical drug discovery. With 13+ years of expertise in mouse model engineering and >500 engineered mouse models under our belt, we can engineer advanced, physiologically relevant mouse models with a wide range of precision genetic modifications specific to your projects’ needs.
We also offer a repository of off-shelf mouse models that can be directly used for studies on gene function, drug screening, and human diseases, thus effectively shortening your overall experimental cycle.

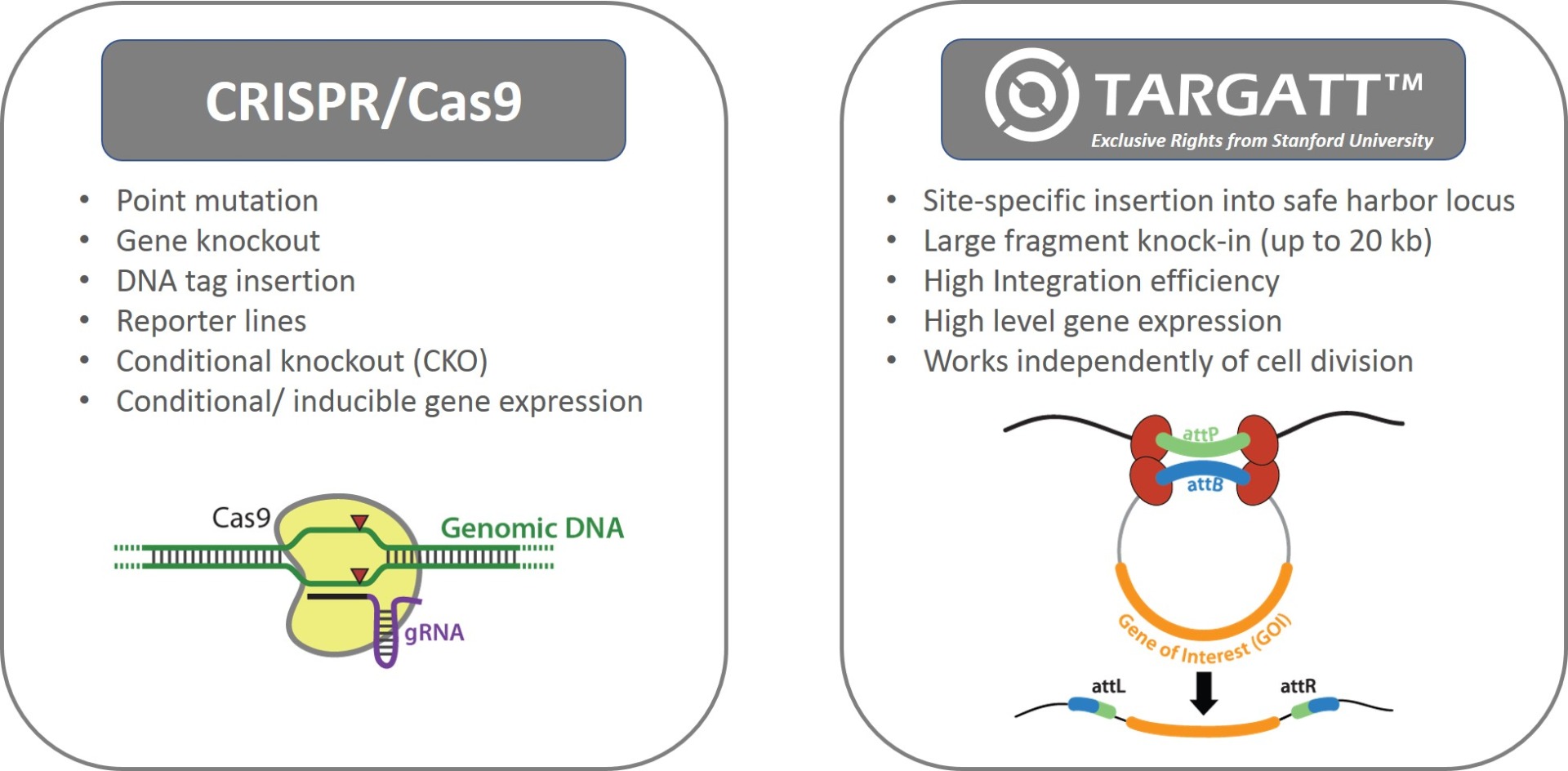
Applied StemCell uses two complementary genome editing technologies to generate advanced cell lines, very efficiently and effectively: the CRISPR/Cas9 technology and our propriety site-specific gene integration technology, TARGATT™ for large fragment (up to 20 kb) knock-in into a safe harbor locus.
|
Project Purpose |
CRISPR/Cas9 |
|
|
Knock-Out (KO) |
Yes |
|
|
Point Mutation |
Yes |
|
|
Conditional KO |
Yes |
|
|
Knock-In (<200 Nucleotide ssODN Donor) |
Yes |
|
|
Knock-In Transgenes in Safe Harbor Loci (>2kb) |
Challenging (but limitations on size) |
Yes (up to 22kb) |
|
Knock-In (Plasmid DNA) |
Challenging (but limitations on size) |
Yes (2 steps: KI docking site; KI transgene) |
Applications: Functional genomics, disease modeling, target identification and validation for drug discovery and screening, and many more.
We also offer mouse model generation service using an expanded technology portfolio such as traditional homologous recombination via ESCs, bacterial artificial chromosome and random transgenic technologies. With our expertise in mouse model generation service and various genome editing technologies, we can assure you a custom genetically engineered mouse model perfect for your research needs.
Homologous Recombination Conditional Knockout & Knock-in Mouse Models
Transgenic Mouse Models, Bacterial Artificial Chromosome & Random Microinjection
Applied StemCell (ASC) also offers ready-for-research genetically engineered mouse models (GEMM) in five categories which are designed for cutting-edge biomedical research and development of human therapeutics:
We also offer downstream phenotyping and drug screening services to accelerate your drug pipeline through early-stage preclinical phases.
CRISPR Technology
Smalley, E. (2016). CRISPR mouse model boom, rat model renaissance. Nature Biotechnology. 34, 893–894.
Baker, M. (2014). Gene editing at CRISPR speed. Nature biotechnology, 32(4), 309-313.
CRISPR Knock-in H11 Locus in Pigs
Ruan, J., Li, H., Xu, K., Wu, T., Wei, J., Zhou, R., ... & Chen-Tsai, R. Y. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Scientific reports, 5, 14253.
Knock-in, Knockout, Conditional Knock-out
*Peng, L., Zhang, H., Hao, Y., Xu, F., Yang, J., Zhang, R., ... & Chen, C. (2016). Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine, 14, 83-96.
*Hu, J. K., Crampton, J. C., Locci, M., & Crotty, S. (2016). CRISPR-mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ triple gene disruption reveals NKT cell defects but not T follicular helper cell defects. PloS one, 11(5), e0156074.
*Besschetnova, T. Y., Ichimura, T., Katebi, N., Croix, B. S., Bonventre, J. V., & Olsen, B. R. (2015). Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biology, 42, 56-73.
*McKenzie, C. W., Craige, B., Kroeger, T. V., Finn, R., Wyatt, T. A., Sisson, J. H., ... & Lee, L. (2015). CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Molecular biology of the cell, 26(18), 3140-3149.
*Bishop, K. A., Harrington, A., Kouranova, E., Weinstein, E. J., Rosen, C. J., Cui, X., & Liaw, L. (2016). CRISPR/Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3: Genes, Genomes, Genetics, 6(7), 2051-2061.
Zhao, M., Tao, F., Venkatraman, A., Li, Z., Smith, S. E., Unruh, J., ... & Marshall, H. (2019). N-Cadherin-Expressing Bone and Marrow Stromal Progenitor Cells Maintain Reserve Hematopoietic Stem Cells. Cell reports, 26(3), 652-669.
Li, C., Zheng, Z., Ha, P., Chen, X., Jiang, W., Sun, S., ... & Chen, E. C. (2018). Neurexin Superfamily Cell Membrane Receptor Contactin‐Associated Protein Like‐4 (Cntnap4) is Involved in Neural EGFL Like 1 (Nell‐1)‐responsive Osteogenesis. Journal of Bone and Mineral Research https://doi.org/10.1002/jbmr.3524.
Geraets, R. D., Langin, L. M., Cain, J. T., Parker, C. M., Beraldi, R., Kovacs, A. D., ... & Pearce, D. A. (2017). A tailored mouse model of CLN2 disease: A nonsense mutant for testing personalized therapies. PloS one, 12(5), e0176526.
Miller, J. N., Kovács, A. D., & Pearce, D. A. (2015). The novel Cln1R151Xmouse model of infantile neuronal ceroid lipofuscinosis (INCL) for testing nonsense suppression therapy. Human Molecular Genetics, 24(1), 185–196. http://doi.org/10.1093/hmg/ddu428.