Newsletter
High-throughput CRISPR iPSC Genome Editing Service
Applied StemCell (ASC) has provided stem cell and genome editing services for over 13 years, and we have worked with researchers all across the globe to engineer over 500 unique cell line models. As one of the earliest providers of CRISPR/Cas9 genome editing services, ASC has the experience and optimized protocols for Rapid Automated Cell Line Editing (RACE™) in induced pluripotent stem cells (iPSCs)!
With our well-established high-throughput protocols, ASC's experts can produce any complex or mainstream genetic modification in your healthy or diseased iPSCs for your basic research, disease modeling, tissue engineering, regenerative medicine, or cell-based therapy research. Leverage our efficient CRISPR genome editing method to obtain your CRISPR-engineered iPS cells in just a few weeks.
-
High success rate: >98% projects completed to customer’s specifications
-
ASC can genetically modify your healthy or diseased iPSCs; control lines are available
-
Single cell cloning (clonal isolation)
-
Homozygous or Heterozygous
-
Automated processes for consistency and high throughput scalability
- Optimized CRISPR iPSC protocols
-
Pluripotency maintained throughout genome editing process using high-end cell culture reagents and protocols
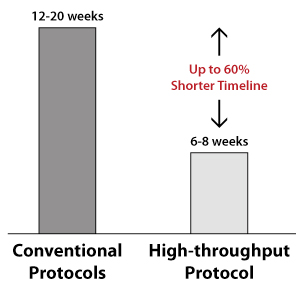
- Fast turnaround time: 6-8 weeks when you select one of the ASC control lines, 3-4 months when you send in your iPSCs
- GMP iPSC Gene Editing Available >> Learn More
ASC is a one-stop-shop for all your iPSC service needs. We are one of the few providers of integrated upstream iPSC generation & downstream differentiation and assay development services. If you are looking to engineer iPSCs in a GMP setting, we invite you to read more about our new GMP-grade iPSC service offerings.
Applied StemCell's CRISPR iPSC Genome Editing Service Compared to Other Providers
| ASC | Other Providers | |
| Final Deliverables | Isogenic Clones with Detailed QC | Mostly pool |
| Choice of Zygosity | Flexible | Fixed Standard |
| Types of Gene Modifications | Flexible | Fixed Standard |
| Starting iPSC Lines | Customer’s, ASC’s Master Lines | Mostly Customer’s only |
| Donor Genetic Background | Normal, Disease-specific | Limited to Healthy |
| QC for Final Product | Standard, Flexible | Mostly Standard |
| Downstream Assays | Available | Mostly Limited |
| Timeline for Projects | 2-3 months | 5-6 months |
Products and Services
Technical Details
Automated High-throughput Protocols for the Genetic Modification of Your iPSC Line of Choice
We can engineer your control and disease iPSC lines or choose from one of our well-characterized master iPSC lines derived from fibroblasts (male: ASE-9211; female: ASE-9209), cord blood, or PBMCs, which have proven CRISPR gene editing and differentiation potential.
|
Faster Timelines with Automated High-throughput Protocols |
|||
|
Project type |
Conventional Protocols |
ASC’s Optimized High-throughput Protocols |
Improvement in Delivery Times |
|
Knockout (KO) |
12-20 weeks |
6-8 weeks |
60% |
|
Point Mutation (Single Nucleotide Polymorphism or Variant) |
12-20 weeks |
6-8 weeks |
60% |
|
Knock-in (Reporters/ Tags, Large Transgenes) |
12-24 weeks |
10-15 weeks |
20-30% |
A Variety of Other Modifications: Standard & Complex
Correct/engineer mutations or introduce a variety of genetic modifications in iPSCs:
- Gene knockout: gene disruption or site-specific large fragment knockout (>10kb)
- Gene insertion: reporter gene/ tag insertion, small fragment insertions, SNV/ point mutations
- Inducible gene expression/ gene overexpression models
- Gene fusion (translocation, inversion, etc)
Don't limit yourself to only the standard modifications. Ask us about: Multiplexed genome editing; conditional knock-in, gene fusion, and other models that you would like for your projects.
And…. We offer Customized Deliverables
- Choice of heterozygous or homozygous mutations
- Footprint-free genome editing – Ex. Single nucleotide variant (SNV; point mutation) engineering without silent mutations for regulatory compliance
- Specific genetic or safe harbor locus
Case Studies
CRISPR Knock-In Projects
Project 1:
Goal: Knock-in of 1 bp at the AAVS1 locus using the ASE-9211 Master iSPC Line by CRISPR/Cas9 technology
Knock-In Strategy for AAVS1 (1bp insertion)
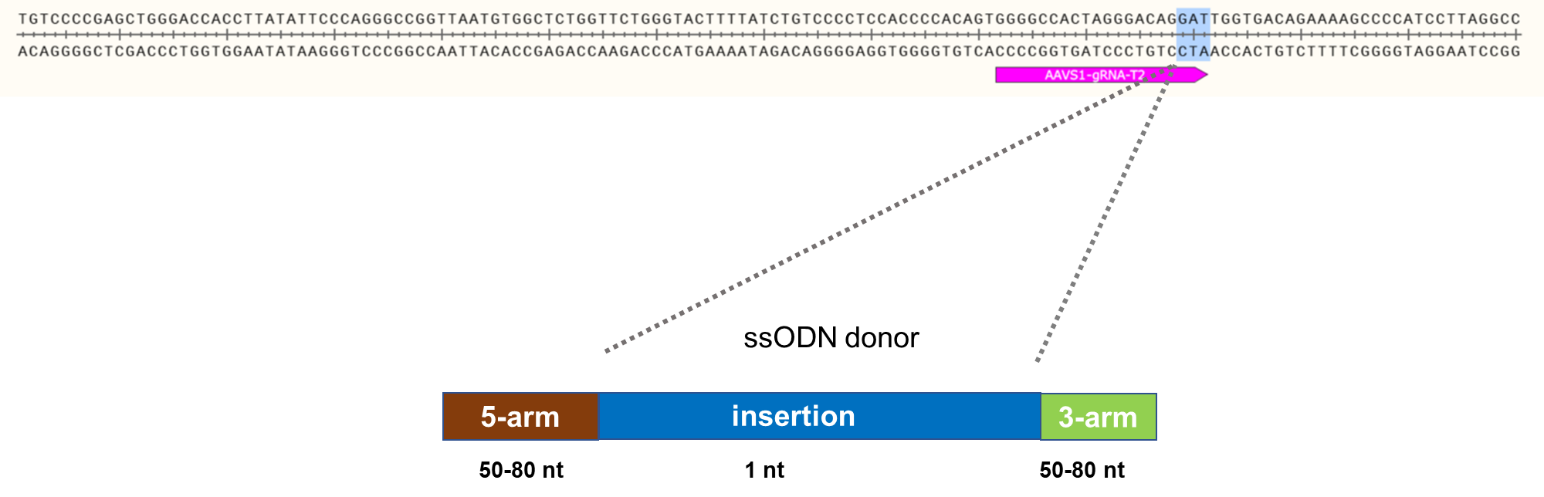
Figure 1: Knock-in strategy for 1bp insertion in the AAVS1 locus of the ASE-9211 Master Cell Line.
Genotyping Clone #6
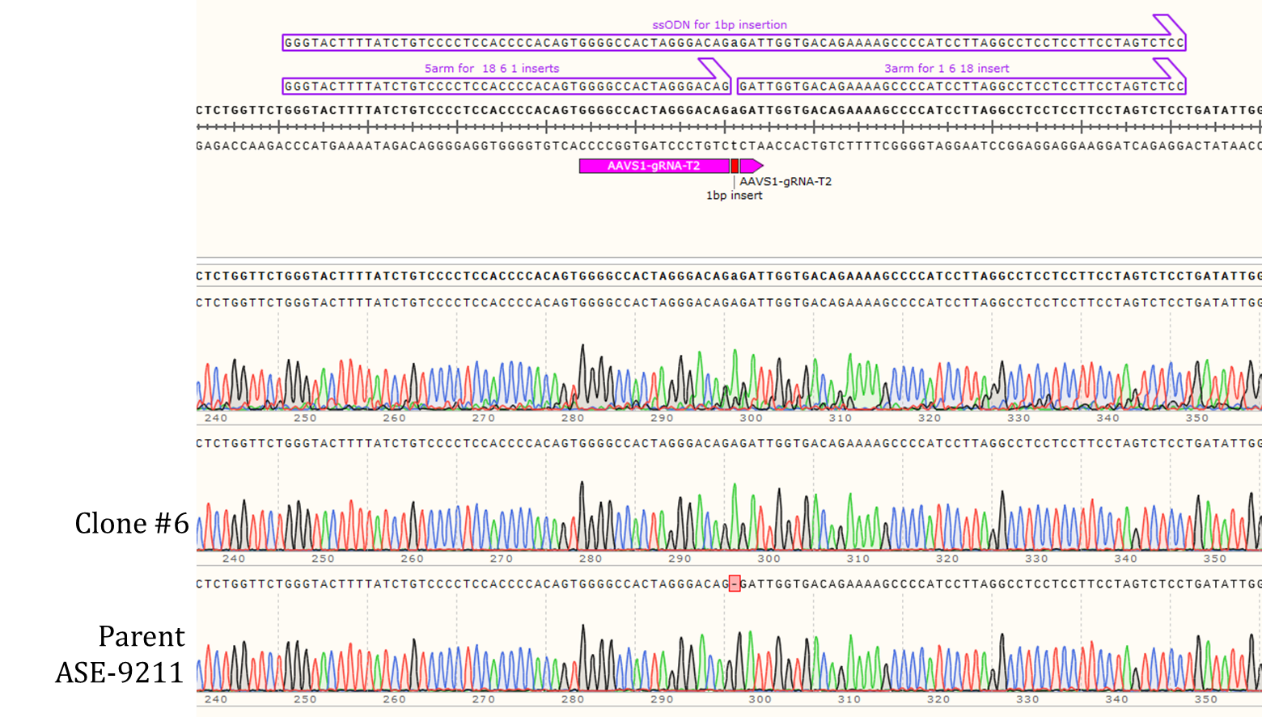
Figure 2: Sequencing chromatogram of iPSC line with 1bp insertion in the AAVS1 locus (top: Clone #6) compared to the Parent line, ASE-9211 (bottom).
Project 2:
Goal: Knock-in of 150bp at the AAVS1 locus using the ASE-9211 Master iPSC Line by CRISPR/Cas9 technology
Knock-In Strategy for AAVS1 (150bp insertion)
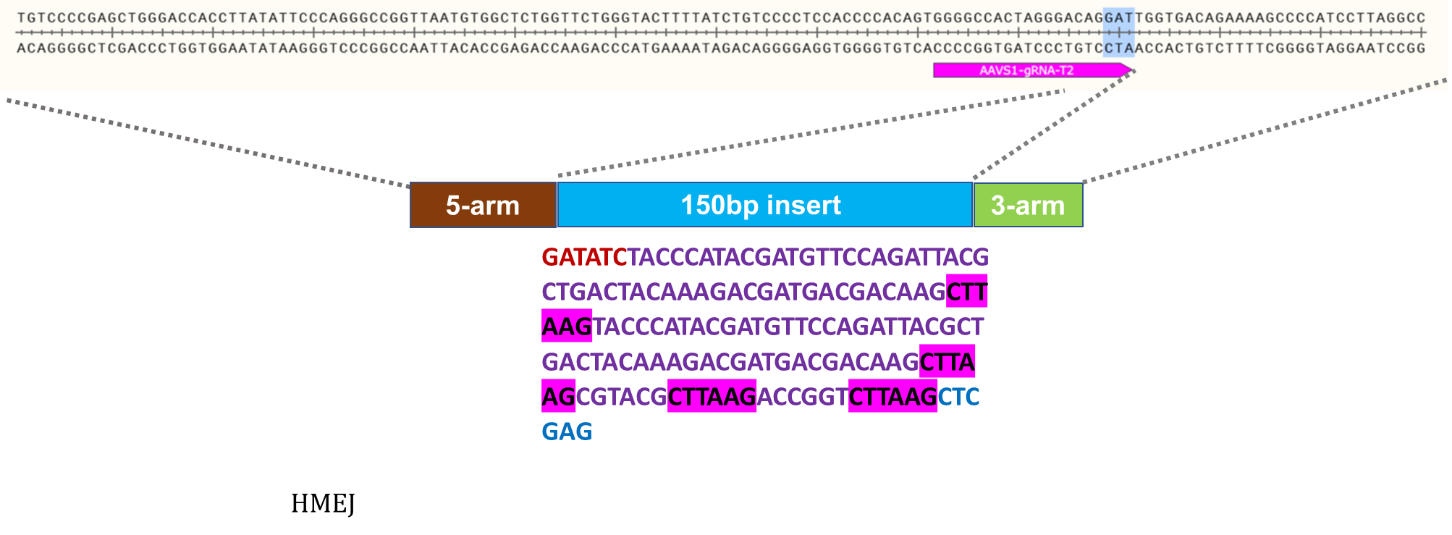
Figure 3: Knock-in strategy for 150bp insertion at the AAVS1 locus of the Master iPSC Line.
Genotyping Positive Clone #21
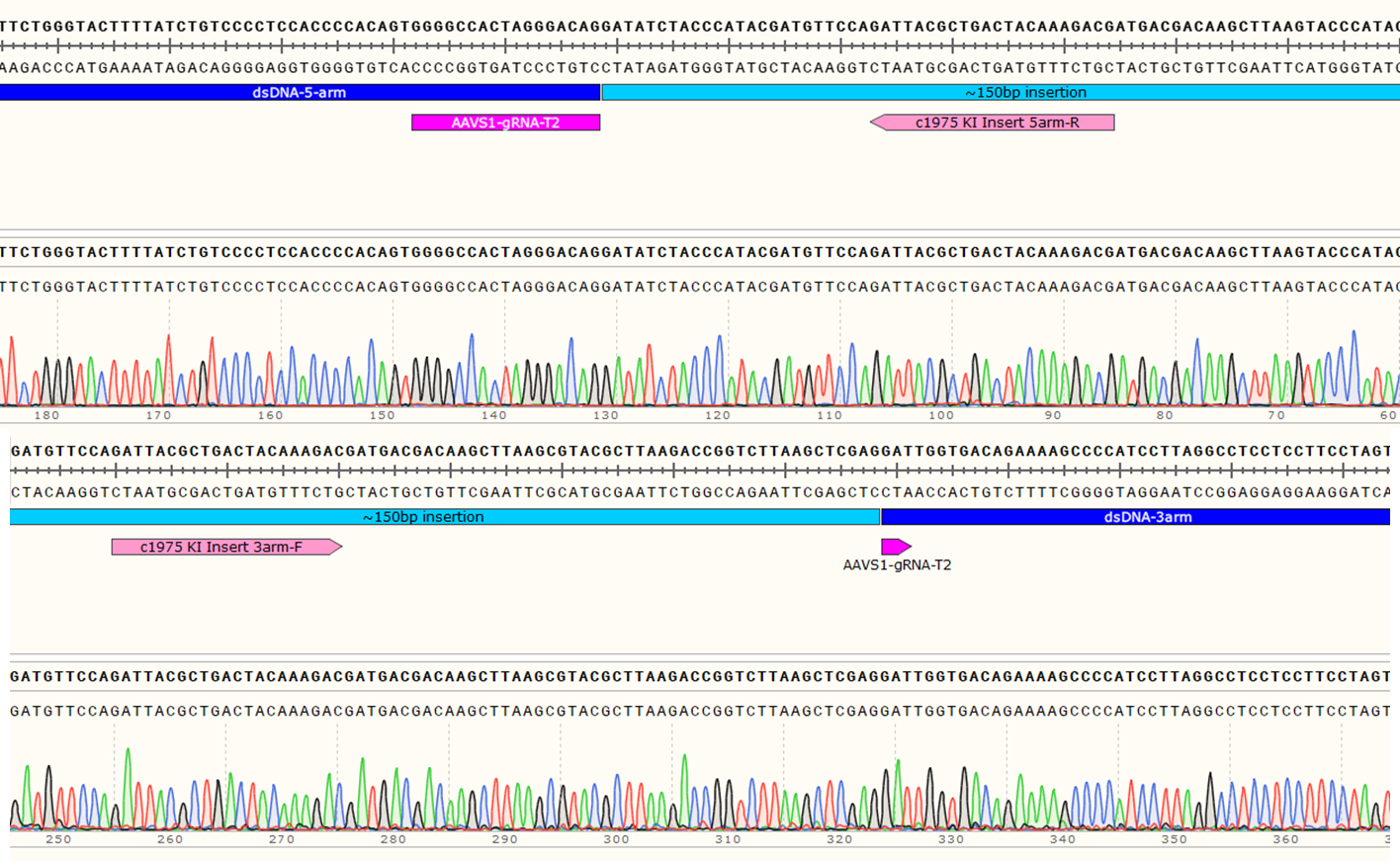
Figure 4: Sequencing chromatogram showing the ~150bp insertion at AAVS1 locus.
CRISPR Knockout Projects
Project 3:
Goal: 1bp deletion in the AAVS1 locus using the ASE-9211 Master Cell Line by CRISPR/Cas9 technology
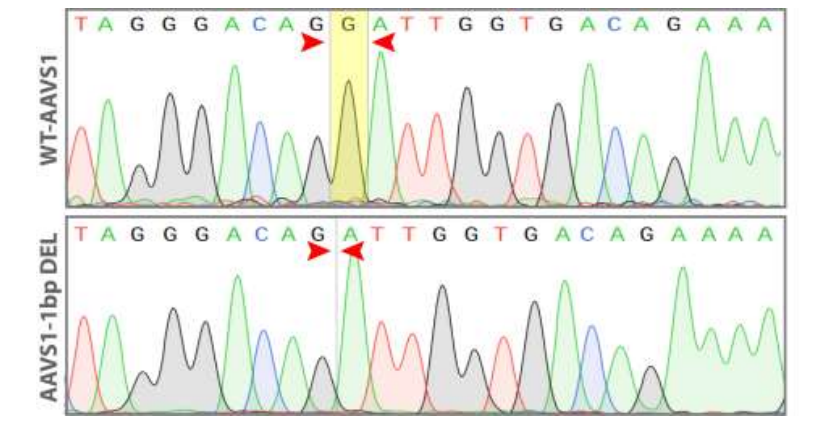
Figure 5. Sequence chromatogram of iPSC line with 1 bp deletion (AAVS1-1bp DEL; bottom) compared to wild type (WT; top).

Figure 6. Sequence alignment between the 1 bp deletion iPSC line (AAVS1-1bp DEL; bottom) and wild type (WT; top).
Project 4:
Goal: 1000bp Deletion in the AAVS1 locus using the ASE-9211 Master Cell Line by CRISPR/Cas9 technology
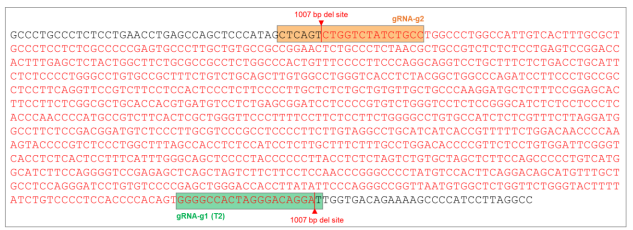
Figure 7. AAVS1 wild type (WT) sequence showing gRNA cut sites and position of 1007 bp (~1000 bp) deletion (sequence in red).
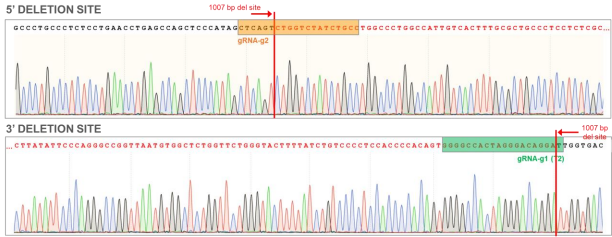
Figure 8. AAVS1 WT chromatogram showing sites of ~1000 bp deletion (sequence in red). Top: Sequence for 5’ deletion site; Bottom: Sequence for 3’ deletion site.
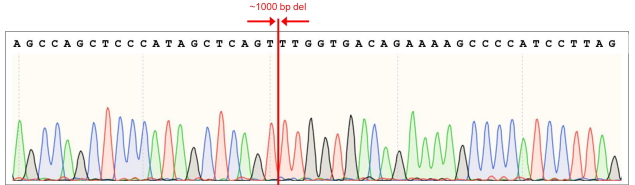
Figure 9. Sequence chromatogram of iPSC line with ~1000 bp deletion in the AAVS1 locus.
Only a few NIST projects are listed, if you would like to learn more, contact us today.
Support Materials
Publications
- Ilic, D. (2019). Latest developments in the field of stem cell research and regenerative medicine compiled from publicly available information and press releases from nonacademic institutions in October 2018. Regenerative medicine, 14(2), 85-92.
- Simkin, D., Searl, T. J., Piyevsky, B. N., Forrest, M., Williams, L. A., Joshi, V., ... & Penzes, P. (2019). Impaired M-current in KCNQ2 Encephalopathy Evokes Dyshomeostatic Modulation of Excitability. bioRxiv, 538371. https://doi.org/10.1101/538371
- Jang, Y., Choi, J., Park, N., Kang, J., Kim, M., Kim, Y., & Ju, J. H. (2019). Development of immunocompatible pluripotent stem cells via CRISPR-based human leukocyte antigen engineering. Experimental & Molecular Medicine, 51(1), 3.
- Lizarraga, S. B., Maguire, A. M., Ma, L., van Dyck, L. I., Wu, Q., Nagda, D., ... & Cowen, M. H. (2018). Human neurons from Christianson syndrome iPSCs reveal allele-specific responses to rescue strategies. bioRxiv, 444232.
- Tanaka, H., Kondo, K., Chen, X., Homma, H., Tagawa, K., Kerever, A., ... & Fujita, K. (2018). The intellectual disability gene PQBP1 rescues Alzheimer’s disease pathology. Molecular Psychiatry, 1.
-
Selvan N., George, S., Serajee, F. J., Shaw, M., Hobson, L., Kalscheuer, V. M., ... & Schwartz, C. E. (2018). O-GlcNAc transferase missense mutations linked to X-linked intellectual disability deregulate genes involved in cell fate determination and signaling. Journal of Biological Chemistry, jbc-RA118.
-
Chai, S., Wan, X., Ramirez-Navarro, A., Tesar, P. J., Kaufman, E. S., Ficker, E., ... & Deschênes, I. (2018). Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. The Journal of clinical investigation, 128(3).
-
Seigel, G. M., et al. (2014). Comparative Analysis of ABCG2+ Stem-Like Retinoblastoma Cells and Induced Pluripotent Stem Cells as Three-Dimensional Aggregates. Investigative Ophthalmology & Visual Science, 55(13), 3068-3068.
-
Comley, J. (2016). CRISPR/Cas9 - transforming gene editing in drug discovery labs. Drug Discovery Weekly. Fall 2016; 33-48.
FAQs
Is the gene editing process feeder independent?
Can you genetically modify my iPSCs?
Can you correct mutations in the iPSCs?
How do you confirm the gene is correctly edited in the iPSCs?




