Newsletter
Rat Models
Genetically engineered rat models are gaining popularity as the preferred biological model for several research areas. ASC can genetically engineer rat models with physiologically relevant modifications using an expanded technology portfolio with CRISPR/Cas9 and traditional homologous recombination and random transgenic technologies.
- High-quality service
- F1 breeding for germline transmission
- New! surgically/ chemically induced rat models of neurological diseases
- Customized projects for in vivo assessments (automated behavior/ locomotor activity, EEG/ ECG, and pharmacokinetics) as well as in vitro evaluations (electrophysiology, immunohistochemistry & other biochemical assays)
Products and Services
Technical Details
For decades, mice have been the major transgenic in vivo model for gene function studies and drug discovery. Their low maintenance cost and easy manipulation made them a preferred tool for pre-clinical research. However, there is still a significant void in animal models required for specific research purposes which the mouse models have been unable to fill. As the need for a better understanding of human genetics has grown, there has been an increased demand for improved transgenic animal models.
Rat models are making a comeback as a preferred in vivo model for researchers who seek a better representation of human genetics and physiology. The successful isolation of rat embryonic stem cells (rES) and complementary accomplishments in site-directed mutagenesis using techniques such as CRISPR/Cas9, ZFNs, and TALENs have made the generation of transgenic rat models possible.
Why do we need transgenic rat models?
- Mouse models are not always reliable in preclinical studies and have several limitations for modeling human diseases
- The metabolism, physiology and pathology of rats are closer to human than mice
- The larger size of rats allows for sophisticated surgeries, instrumentation and manipulations
- Excellent behavioral models for cognition and memory, neurological and psychological assays and drug screening
- Superior model for cardiovascular diseases (stroke and hypertension), autoimmune disorders, diabetes, breast cancer, and autoimmune disorders
Comparison of Transgenic Technologies
|
Methods |
Technical Advantage |
|
CRISPR / Cas9 |
|
Some examples of available genetic modifications in rats include but not limited to:
- Constitutive and conditional knockouts
- Small fragment insertions, point mutations
- Large fragment knock-in
- Gene tagging/ reporter gene insertion
- Gene replacement
- Gene fusion/ translocation
- Gene overexpression, inducible expression, promoter modifications
- Gene editing/ correction to model human diseases
Applications: Functional genomics, disease modeling, target identification and validation for drug discovery and screening, and many more.
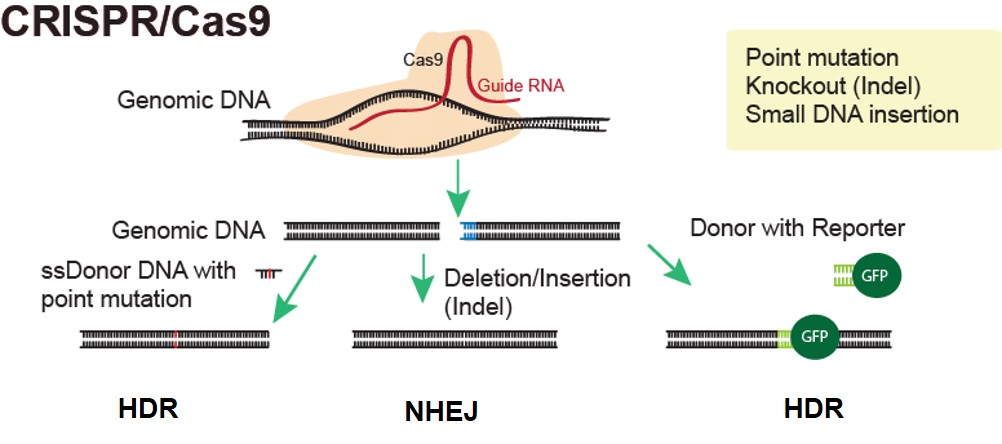
Applied StemCell has worked with CRISPR to generate advanced cell lines and animal models very efficiently and effectively.
|
Project Purpose |
CRISPR/Cas9 |
|
Knock-Out (KO) |
Yes |
|
Point Mutation |
Yes |
|
Conditional KO |
Yes |
|
Knock-In (<200 Nucleotide ssODN Donor) |
Yes |
|
Knock-In Transgenes in Safe Harbor Loci (>2kb) |
Challenging (but limitations on size) |
|
Knock-In (Plasmid DNA) |
Challenging (but limitations on size) |
We currently offer mouse model generation services using an expanded technology portfolio such as traditional homologous recombination via ESCs, bacterial artificial chromosomes, and random transgenic technologies. With our expertise in mouse model generation service and various genome editing technologies, we can assure you a custom genetically engineered mouse model perfect for your research needs.
Support Materials
Publications
CRISPR Mouse/ Rat Models: Knock-in, Knockout, and Conditional Knockout
CRISPR Technology
- Smalley, E. (2016). CRISPR mouse model boom, rat model renaissance. Nature Biotechnology. 34, 893–894.
- Baker, M. (2014). Gene editing at CRISPR speed. Nature biotechnology, 32(4), 309-313.
CRISPR Knock-in H11 Locus in Pigs
- Ruan, J., Li, H., Xu, K., Wu, T., Wei, J., Zhou, R., ... & Chen-Tsai, R. Y. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Scientific reports, 5, 14253.
Knock-in, Knockout, Conditional Knock-out
- Park, J., Jung, E., Lee, S. H., & Chung, W. S. (2020). CDC50A dependent phosphatidylserine exposure induces inhibitory post-synapse elimination by microglia. bioRxiv.
- Ramachandra Rao, S., Fliesler, S. J., Kotla, P., Nguyen, M. N., & Pittler, S. J. (2020). Lack of Overt Retinal Degeneration in a K42E Dhdds Knock-In Mouse Model of RP59. Cells, 9(4), 896.
- Beurg, M., Barlow, A., Furness, D. N., & Fettiplace, R. (2019). A Tmc1 mutation reduces calcium permeability and expression of mechanoelectrical transduction channels in cochlear hair cells. Proceedings of the National Academy of Sciences, 116(41), 20743-20749.
- Goldring, A. C., Beurg, M., & Fettiplace, R. (2019). The contribution of TMC1 to adaptation of mechanoelectrical transduction channels in cochlear outer hair cells. The Journal of physiology.
- Hwang, S., He, Y., Xiang, X., Seo, W., Kim, S. J., Ma, J., ... & Kunos, G. (2019). Interleukin‐22 ameliorates neutrophil‐driven nonalcoholic steatohepatitis through multiple targets. Hepatology https://doi.org/10.1002/hep.31031.
- Dumesic, P. A., Egan, D. F., Gut, P., Tran, M. T., Parisi, A., Chatterjee, N., ... & Dou, F. (2019). An Evolutionarily Conserved uORF Regulates PGC1α and Oxidative Metabolism in Mice, Flies, and Bluefin Tuna. Cell metabolism.
- Liang, T., Zhang, H., Xu, Q., Wang, S., Qin, C., & Lu, Y. (2019). Mutant Dentin Sialophosphoprotein Causes Dentinogenesis Imperfecta. Journal of dental research, 0022034519854029.
- Qian, W., Miner, C. A., Ingle, H., Platt, D. J., Baldridge, M. T., & Miner, J. J. (2019). A human STAT1 gain-of-function mutation impairs CD8+ T cell responses against gammaherpesvirus-68. Journal of virology, JVI-00307.
- Kweon, S. M., Chen, Y., Moon, E., Kvederaviciutė, K., Klimasauskas, S., & Feldman, D. E. (2019). An Adversarial DNA N6-Methyladenine-Sensor Network Preserves Polycomb Silencing. Molecular Cell. https://doi.org/10.1016/j.molcel.2019.03.018
- Deng, F., He, S., Cui, S., Shi, Y., Tan, Y., Li, Z., ... & Peng, L. (2018). A Molecular Targeted Immunotherapeutic Strategy for Ulcerative Colitis via Dual-Targeting Nanoparticles Delivering miR-146b to Intestinal Macrophages. Journal of Crohn's and Colitis.
- Jo, S., Fonseca, T. L., Bocco, B. M. D. C., Fernandes, G. W., McAninch, E. A., Bolin, A. P., ... & Németh, D. (2018). Type 2 deiodinase polymorphism causes ER stress and hypothyroidism in the brain. The Journal of Clinical Investigation.
- Langston, R. G., Rudenko, I. N., Kumaran, R., Hauser, D. N., Kaganovich, A., Ponce, L. B., ... & Beilina, A. (2018). Differences in Stability, Activity and Mutation Effects Between Human and Mouse Leucine-Rich Repeat Kinase 2. Neurochemical research, 1-14.
- Amara, N., Tholen, M., & Bogyo, M. (2018). Chemical tools for selective activity profiling of endogenously expressed MMP-14 in multicellular models. ACS Chemical Biology. doi: 10.1021/acschembio.8b00562.
- Allocca, S., Ciano, M., Ciardulli, M. C., D’Ambrosio, C., Scaloni, A., Sarnataro, D., ... & Bonatti, S. (2018). An αB-Crystallin Peptide Rescues Compartmentalization and Trafficking Response to Cu Overload of ATP7B-H1069Q, the Most Frequent Cause of Wilson Disease in the Caucasian Population. International journal of molecular sciences, 19(7).
- Peng, L., Zhang, H., Hao, Y., Xu, F., Yang, J., Zhang, R., ... & Chen, C. (2016). Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine, 14, 83-96.
- Hu, J. K., Crampton, J. C., Locci, M., & Crotty, S. (2016). CRISPR-mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ triple gene disruption reveals NKT cell defects but not T follicular helper cell defects. PloS one, 11(5), e0156074.
- Besschetnova, T. Y., Ichimura, T., Katebi, N., Croix, B. S., Bonventre, J. V., & Olsen, B. R. (2015). Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biology, 42, 56-73.
- McKenzie, C. W., Craige, B., Kroeger, T. V., Finn, R., Wyatt, T. A., Sisson, J. H., ... & Lee, L. (2015). CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Molecular biology of the cell, 26(18), 3140-3149.
- Bishop, K. A., Harrington, A., Kouranova, E., Weinstein, E. J., Rosen, C. J., Cui, X., & Liaw, L. (2016). CRISPR/Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3: Genes, Genomes, Genetics, 6(7), 2051-2061.
TARGATT™ Site Specific Knock-in Mouse
Book Chapters
- Chen-Tsai, R. Y. (2020). Integrase-Mediated Targeted Transgenics Through Pronuclear Microinjection. In Transgenic Mouse (pp. 35-46). Humana, New York, NY.
- Chen-Tsai, R. Y. (2019). Using TARGATT™ Technology to Generate Site-Specific Transgenic Mice. In Microinjection (pp. 71-86). Humana Press, New York, NY.
Master Cell Line
- Chi, X., Zheng, Q., Jiang, R., Chen-Tsai, R. Y., & Kong, L. J. (2019). A system for site-specific integration of transgenes in mammalian cells. PLOS ONE, 14(7), e0219842.
Description of the technology
- Zhu, F., Gamboa, M., Farruggio, A. P., Hippenmeyer, S., Tasic, B., Schüle, B., … Calos, M. P. (2014). DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Research, 42(5), e34. http://doi.org/10.1093/nar/gkt1290.
- Tasic, B., Hippenmeyer, S., Wang, C., Gamboa, M., Zong, H., Chen-Tsai, Y., & Luo, L. (2011). Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proceedings of the National Academy of Sciences of the United States of America, 108(19), 7902–7907. http://doi.org/10.1073/pnas.1019507108.
Commentary, comparison with other transgenic methods
- Rossant, J., Nutter, L. M., & Gertsenstein, M. (2011). Engineering the embryo. Proceedings of the National Academy of Sciences, 108(19), 7659-7660.
Tet inducible mice generated by TARGATT™
- Fan, X., Petitt, M., Gamboa, M., Huang, M., Dhal, S., Druzin, M. L., … Nayak, N. R. (2012). Transient, Inducible, Placenta-Specific Gene Expression in Mice. Endocrinology, 153(11), 5637–5644. http://doi.org/10.1210/en.2012-1556.
Advantage of Hipp11 (H11) locus
- Hippenmeyer, S., Youn, Y. H., Moon, H. M., Miyamichi, K., Zong, H., Wynshaw-Boris, A., & Luo, L. (2010). Genetic Mosaic Dissection of Lis1 and Ndel1 in Neuronal Migration. Neuron, 68(4), 695–709. http://doi.org/10.1016/j.neuron.2010.09.027.
Applications for mice generated by TARGATT™ (and cited/published articles)
- Lindtner, S., Catta-Preta, R., Tian, H., Su-Feher, L., Price, J. D., Dickel, D. E., ... & Pennacchio, L. A. (2019). Genomic Resolution of DLX-Orchestrated Transcriptional Circuits Driving Development of Forebrain GABAergic Neurons. Cell reports, 28(8), 2048-2063.
- Wang, T. A., Teo, C. F., Åkerblom, M., Chen, C., Tynan-La Fontaine, M., Greiner, V. J., ... & Jan, L. Y. (2019). Thermoregulation via Temperature-Dependent PGD2 Production in Mouse Preoptic Area. Neuron, 103(2), 309-322.
- Clarke, B. A., Majumder, S., Zhu, H., Lee, Y. T., Kono, M., Li, C., ... & Byrnes, C. (2019). The Ormdl genes regulate the sphingolipid synthesis pathway to ensure proper myelination and neurologic function in mice. eLife, 8.
- Carlson, H. L., & Stadler, H. S. (2019). Development and functional characterization of a lncRNA‐HIT conditional loss of function allele. genesis, e23351.
- Chande, S., Ho, B., Fetene, J., & Bergwitz, C. (2019). Transgenic mouse model for conditional expression of influenza hemagglutinin-tagged human SLC20A1/PIT1. PloS one, 14(10), e0223052. doi:10.1371/journal.pone.0223052
- Hu, Q., Ye, Y., Chan, L. C., Li, Y., Liang, K., Lin, A., ... & Pan, Y. (2019). Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nature immunology, 1.
- Matharu, N., Rattanasopha, S., Tamura, S., Maliskova, L., Wang, Y., Bernard, A., ... & Ahituv, N. (2018). CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science, eaau0629.
- Barrett, R. D., Laurent, S., Mallarino, R., Pfeifer, S. P., Xu, C. C., Foll, M., ... & Hoekstra, H. E. (2018). The fitness consequences of genetic variation in wild populations of mice. bioRxiv, 383240.
- Ibrahim, L. A., Huang, J. J., Wang, S. Z., Kim, Y. J., Li, I., & Huizhong, W. (2018). Sparse Labeling and Neural Tracing in Brain Circuits by STARS Strategy: Revealing Morphological Development of Type II Spiral Ganglion Neurons. Cerebral Cortex, 1-14.
- Kumar, A., Dhar, S., Campanelli, G., Butt, N. A., Schallheim, J. M., Gomez, C. R., & Levenson, A. S. (2018). MTA 1 drives malignant progression and bone metastasis in prostate cancer. Molecular oncology.
- Jang, Y., Wang, C., Broun, A., Park, Y. K., Zhuang, L., Lee, J. E., ... & Ge, K. (2018). H3. 3K4M destabilizes enhancer epigenomic writers MLL3/4 and impairs adipose tissue development. bioRxiv, 301986. doi:https://doi.org/10.1101/301986
- Tang, Y., Kwon, H., Neel, B. A., Kasher-Meron, M., Pessin, J., Yamada, E., & Pessin, J. E. (2018). The fructose-2, 6-bisphosphatase TIGAR suppresses NF-κB signaling by directly inhibiting the linear ubiquitin assembly complex LUBAC. Journal of Biological Chemistry, jbc-RA118.
- Chen, M., Geoffroy, C. G., Meves, J. M., Narang, A., Li, Y., Nguyen, M. T., ... & Elzière, L. (2018). Leucine Zipper-Bearing Kinase Is a Critical Regulator of Astrocyte Reactivity in the Adult Mammalian CNS. Cell Reports, 22(13), 3587-3597.
- Kido, T., Sun, Z., & Lau, Y.-F. C. (2017). Aberrant activation of the human sex-determining gene in early embryonic development results in postnatal growth retardation and lethality in mice. Scientific Reports, 7, 4113. http://doi.org/10.1038/s41598-017-04117-6.
- Nouri, N., & Awatramani, R. (2017). A novel floor plate boundary defined by adjacent En1 and Dbx1 microdomains distinguishes midbrain dopamine and hypothalamic neurons. Development, 144(5), 916-927.
- Li, K., Wang, F., Cao, W. B., Lv, X. X., Hua, F., Cui, B., ... & Yu, J. M. (2017). TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of p53-Mediated Senescence. Cancer Cell, 31(5), 697-710.
- Matharu, N., Rattanasopha, S., Maliskova, L., Wang, Y., Hardin, A., Vaisse, C., & Ahituv, N. (2017). Promoter or Enhancer Activation by CRISPRa Rescues Haploinsufficiency Caused Obesity. bioRxiv, 140426.
- Jiang, T., Kindt, K., & Wu, D. K. (2017). Transcription factor Emx2 controls stereociliary bundle orientation of sensory hair cells. eLife, 6, e23661.
- Booze, M. L., Hansen, J. M., & Vitiello, P. F. (2016). A Novel Mouse Model for the Identification of Thioredoxin-1 Protein Interactions. Free Radical Biology & Medicine, 99, 533–543. http://doi.org/10.1016/j.freeradbiomed.2016.09.013.
- Feng, D., Dai, S., Liu, F., Ohtake, Y., Zhou, Z., Wang, H., ... & Hayat, U. (2016). Cre-inducible human CD59 mediates rapid cell ablation after intermedilysin administration. The Journal of clinical investigation, 126(6), 2321-2333.
- Sun, N., Yun, J., Liu, J., Malide, D., Liu, C., Rovira, I. I., … Finkel, T. (2015). Measuring in vivo mitophagy. Molecular Cell, 60(4), 685–696. http://doi.org/10.1016/j.molcel.2015.10.009.
- Devine, W. P., Wythe, J. D., George, M., Koshiba-Takeuchi, K., & Bruneau, B. G. (2014). Early patterning and specification of cardiac progenitors in gastrulating mesoderm. eLife, 3, e03848. http://doi.org/10.7554/eLife.03848.
- Fogg, P. C. M., Colloms, S., Rosser, S., Stark, M., & Smith, M. C. M. (2014). New Applications for Phage Integrases. Journal of Molecular Biology, 426(15), 2703–2716. http://doi.org/10.1016/j.jmb.2014.05.014.
- Chen-Tsai, R. Y., Jiang, R., Zhuang, L., Wu, J., Li, L., & Wu, J. (2014). Genome editing and animal models. Chinese science bulletin, 59(1), 1-6.
- Park, K.-E., Park, C.-H., Powell, A., Martin, J., Donovan, D. M., & Telugu, B. P. (2016). Targeted Gene Knockin in Porcine Somatic Cells Using CRISPR/Cas Ribonucleoproteins. International Journal of Molecular Sciences, 17(6), 810. http://doi.org/10.3390/ijms17060810.
- Guenther, C. A., Tasic, B., Luo, L., Bedell, M. A., & Kingsley, D. M. (2014). A molecular basis for classic blond hair color in Europeans. Nature Genetics, 46(7), 748–752. http://doi.org/10.1038/ng.2991.
- Villamizar, C. A. (2014). Characterization of the vascular pathology in the acta2 r258c mouse model and cerebrovascular characterization of the acta2 null mouse. UT GSBS Dissertations and These (Open Access). Paper 508 (2014)
Mouse/ Rat Models: Homologous Recombination Conditional Knockout Mouse
- Geraets, R. D. (2019). Neuronal Ceroid Lipfuscinosis: A Tailored Animal Model of CLN2 Disease and Evaluation of Select Personalized Therapies (Doctoral dissertation, ProQuest Dissertations Publishing).
-
Zhao, M., Tao, F., Venkatraman, A., Li, Z., Smith, S. E., Unruh, J., ... & Marshall, H. (2019). N-Cadherin-Expressing Bone and Marrow Stromal Progenitor Cells Maintain Reserve Hematopoietic Stem Cells. Cell reports, 26(3), 652-669.
-
Li, C., Zheng, Z., Ha, P., Chen, X., Jiang, W., Sun, S., ... & Chen, E. C. (2018). Neurexin Superfamily Cell Membrane Receptor Contactin‐Associated Protein Like‐4 (Cntnap4) is Involved in Neural EGFL Like 1 (Nell‐1)‐responsive Osteogenesis. Journal of Bone and Mineral Research https://doi.org/10.1002/jbmr.3524.
-
Geraets, R. D., Langin, L. M., Cain, J. T., Parker, C. M., Beraldi, R., Kovacs, A. D., ... & Pearce, D. A. (2017). A tailored mouse model of CLN2 disease: A nonsense mutant for testing personalized therapies. PloS one, 12(5), e0176526.
-
Miller, J. N., Kovács, A. D., & Pearce, D. A. (2015). The novel Cln1R151Xmouse model of infantile neuronal ceroid lipofuscinosis (INCL) for testing nonsense suppression therapy. Human Molecular Genetics, 24(1), 185–196. http://doi.org/10.1093/hmg/ddu428.




