Newsletter
PrimeCKO™ Conditional Knockout Rat Models
Applied StemCell can generate custom conditional knockout rat models using CRISPR/Cas9 or other suitable gene-editing techniques to insert the 5’ and 3’ flanking LoxP sequences into the endogenous locus of your gene of interest. These conditional knockout rats can be bred with commercially available Cre expressing rats or custom-engineered Cre expressing rats to generate your rat model with precise control over where and when your gene is knocked out.
ASC offers Bacterial Artificial Chromosomes (BACs) to generate rat models. BACs are ultra-low copy vectors that hold up to 300 kb of genomic fragments.
- High-quality service
- F1 breeding for germline transmission
- Customized projects for in vivo assessments (automated behavior/ locomotor activity, EEG/ ECG, and pharmacokinetics) as well as in vitro evaluations (electrophysiology, immunohistochemistry & other biochemical assays
Products and Services
Technical Details
Gene knockout animal models are extremely important for studying functional importance of genes and the role played by gene dysfunction due to deletions of mutations in specific diseases. While conventional knockout models where gene expression is disrupted at germline level can be a very valuable research tool, constitutive knockout of some critical genes can pose problems such as embryonic lethality, undesired phenotypes and compensatory mechanisms. Conditional Knockout rat models overcome these limitations and offer a refined and versatile approach to study gene dysfunction by limiting the disruption of gene expression to a particular tissue or a specific developmental time of the animal.
One of the most used conditional knockout method is the Cre- LoxP system, in which a gene of interest (targeted exons) is flanked by two 34-bp LoxP sequences (also called floxed alleles) at the desired genomic locus (figure 1). The LoxP sites serve as a target for the Cre-recombinase, which catalyzes deletion of the floxed exon(s) (figure 2).
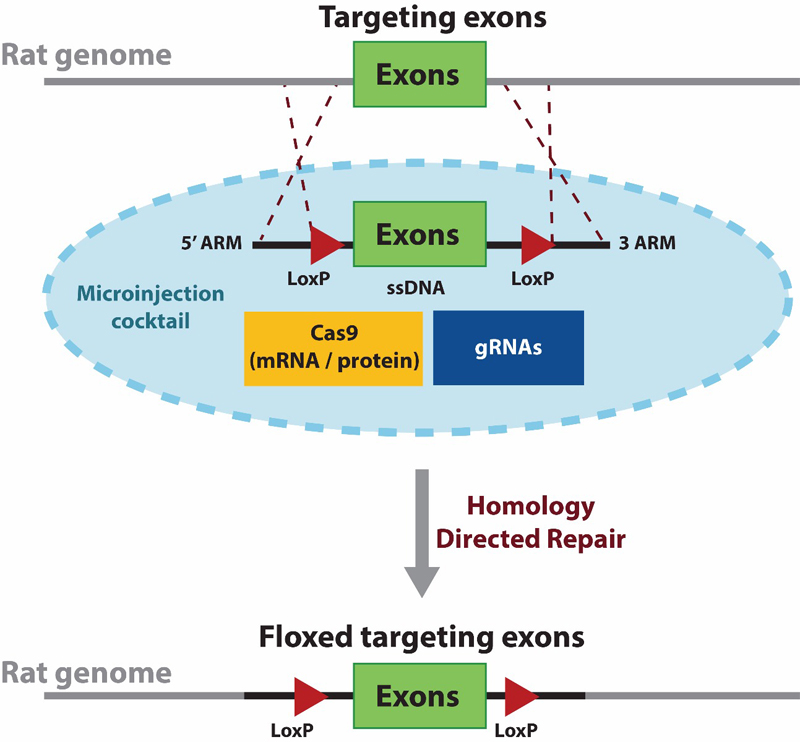
Figure 1. The schematic describes the first stage in developing a conditional knockout rat model CRISPR to generate a floxed (LoxP flanked exon) rat. A single stranded donor DNA (ssDNA) is used for delivering the floxed targeting exons to replace the wildtype form. The donor contains two LoxP sequences flanking the targeted exon(s) along with 5' and 3' homologous arms for directing a site-specific homology directed repair. The donor ssDNA is delivered along with Cas9 (mRNA or protein) and validated gRNAs via microinjection.
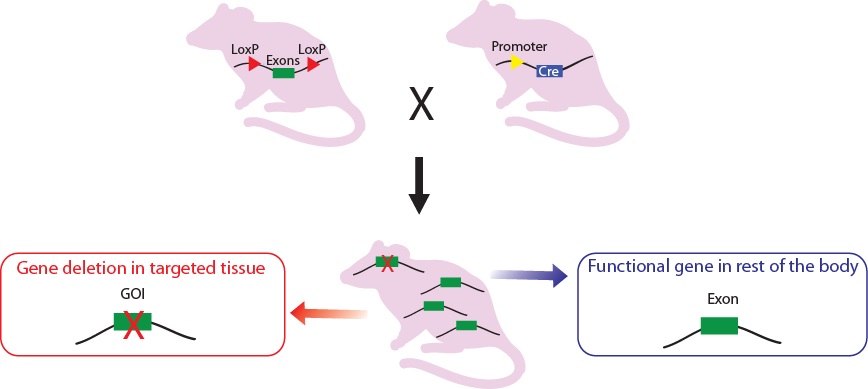
Figure 2. Crossbreeding the conditional knock-out rat with a Cre-recombinase expressing rat. The Cre expression is driven by a promoter of choice: tissue specific or ubiquitous promoter. As an example, a CNS-specific promoter is shown in the figure. The expressed Cre recombinase deletes the floxed exon(s) in a spatial specific manner there by causing a frame shift in downstream sequence.
Looking for a neuronal lineage-specific or other custom Cre Rat Line?
Applied StemCell can generate Cre Rat Lines using its proprietary TARGATT™ technology to suit your research needs. The TARGATT™ integrase-based site-specific gene integration technology is ideal for insertion of large transgenes into a preselected safe harbor locus such as rH11 or rRosa26. Using this technology, the Cre gene under the control of a promoter-of-choice can be inserted into the preselected safe harbor locus for high-level Cre expression in the desired tissue lineage. Some examples of Cre rat lines we can generate are below:
|
Neuronal promoters: |
|
|
Nes |
PAG |
|
CaMK2a |
Syn1 |
|
Wnt1 |
Thy1 |
|
HB8 |
PDGF |
|
MOR23 |
Crh |
|
Pomc |
GFAP |
|
Drd1a |
Six3 |
|
TH |
Plp1 |
|
GAD67 |
Tie2 |
|
Other Promoters: |
|
|
VE-Cad |
|
|
CA |
|
|
CA-LoxP-STOP-LoxP-GFP-LacZ |
|
For other promoters, please inquire.
Support Materials
Publications
CRISPR Mouse/ Rat Models: Knock-in, Knockout, and Conditional Knockout
CRISPR Technology
-
Smalley, E. (2016). CRISPR mouse model boom, rat model renaissance. Nature Biotechnology. 34, 893–894.
-
Baker, M. (2014). Gene editing at CRISPR speed. Nature biotechnology, 32(4), 309-313.
CRISPR Knock-in H11 Locus in Pigs
-
Ruan, J., Li, H., Xu, K., Wu, T., Wei, J., Zhou, R., ... & Chen-Tsai, R. Y. (2015). Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Scientific reports, 5, 14253.
Knock-in, Knockout, Conditional Knock-out
- Park, J., Jung, E., Lee, S. H., & Chung, W. S. (2020). CDC50A dependent phosphatidylserine exposure induces inhibitory post-synapse elimination by microglia. bioRxiv.
- Ramachandra Rao, S., Fliesler, S. J., Kotla, P., Nguyen, M. N., & Pittler, S. J. (2020). Lack of Overt Retinal Degeneration in a K42E Dhdds Knock-In Mouse Model of RP59. Cells, 9(4), 896.
- Beurg, M., Barlow, A., Furness, D. N., & Fettiplace, R. (2019). A Tmc1 mutation reduces calcium permeability and expression of mechanoelectrical transduction channels in cochlear hair cells. Proceedings of the National Academy of Sciences, 116(41), 20743-20749.
- Goldring, A. C., Beurg, M., & Fettiplace, R. (2019). The contribution of TMC1 to adaptation of mechanoelectrical transduction channels in cochlear outer hair cells. The Journal of physiology.
- Hwang, S., He, Y., Xiang, X., Seo, W., Kim, S. J., Ma, J., ... & Kunos, G. (2019). Interleukin‐22 ameliorates neutrophil‐driven nonalcoholic steatohepatitis through multiple targets. Hepatology https://doi.org/10.1002/hep.31031.
- Dumesic, P. A., Egan, D. F., Gut, P., Tran, M. T., Parisi, A., Chatterjee, N., ... & Dou, F. (2019). An Evolutionarily Conserved uORF Regulates PGC1α and Oxidative Metabolism in Mice, Flies, and Bluefin Tuna. Cell metabolism.
- Liang, T., Zhang, H., Xu, Q., Wang, S., Qin, C., & Lu, Y. (2019). Mutant Dentin Sialophosphoprotein Causes Dentinogenesis Imperfecta. Journal of dental research, 0022034519854029.
- Qian, W., Miner, C. A., Ingle, H., Platt, D. J., Baldridge, M. T., & Miner, J. J. (2019). A human STAT1 gain-of-function mutation impairs CD8+ T cell responses against gammaherpesvirus-68. Journal of virology, JVI-00307.
- Kweon, S. M., Chen, Y., Moon, E., Kvederaviciutė, K., Klimasauskas, S., & Feldman, D. E. (2019). An Adversarial DNA N6-Methyladenine-Sensor Network Preserves Polycomb Silencing. Molecular Cell. https://doi.org/10.1016/j.molcel.2019.03.018
- Deng, F., He, S., Cui, S., Shi, Y., Tan, Y., Li, Z., ... & Peng, L. (2018). A Molecular Targeted Immunotherapeutic Strategy for Ulcerative Colitis via Dual-Targeting Nanoparticles Delivering miR-146b to Intestinal Macrophages. Journal of Crohn's and Colitis.
- Jo, S., Fonseca, T. L., Bocco, B. M. D. C., Fernandes, G. W., McAninch, E. A., Bolin, A. P., ... & Németh, D. (2018). Type 2 deiodinase polymorphism causes ER stress and hypothyroidism in the brain. The Journal of Clinical Investigation.
- Langston, R. G., Rudenko, I. N., Kumaran, R., Hauser, D. N., Kaganovich, A., Ponce, L. B., ... & Beilina, A. (2018). Differences in Stability, Activity and Mutation Effects Between Human and Mouse Leucine-Rich Repeat Kinase 2. Neurochemical research, 1-14.
- Amara, N., Tholen, M., & Bogyo, M. (2018). Chemical tools for selective activity profiling of endogenously expressed MMP-14 in multicellular models. ACS Chemical Biology. doi: 10.1021/acschembio.8b00562.
- Allocca, S., Ciano, M., Ciardulli, M. C., D’Ambrosio, C., Scaloni, A., Sarnataro, D., ... & Bonatti, S. (2018). An αB-Crystallin Peptide Rescues Compartmentalization and Trafficking Response to Cu Overload of ATP7B-H1069Q, the Most Frequent Cause of Wilson Disease in the Caucasian Population. International journal of molecular sciences, 19(7).
-
*Peng, L., Zhang, H., Hao, Y., Xu, F., Yang, J., Zhang, R., ... & Chen, C. (2016). Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine, 14, 83-96.
-
*Hu, J. K., Crampton, J. C., Locci, M., & Crotty, S. (2016). CRISPR-mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ triple gene disruption reveals NKT cell defects but not T follicular helper cell defects. PloS one, 11(5), e0156074.
-
*Besschetnova, T. Y., Ichimura, T., Katebi, N., Croix, B. S., Bonventre, J. V., & Olsen, B. R. (2015). Regulatory mechanisms of anthrax toxin receptor 1-dependent vascular and connective tissue homeostasis. Matrix Biology, 42, 56-73.
-
*McKenzie, C. W., Craige, B., Kroeger, T. V., Finn, R., Wyatt, T. A., Sisson, J. H., ... & Lee, L. (2015). CFAP54 is required for proper ciliary motility and assembly of the central pair apparatus in mice. Molecular biology of the cell, 26(18), 3140-3149.
-
*Bishop, K. A., Harrington, A., Kouranova, E., Weinstein, E. J., Rosen, C. J., Cui, X., & Liaw, L. (2016). CRISPR/Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3: Genes, Genomes, Genetics, 6(7), 2051-2061.
Mouse/ Rat Models: Homologous Recombination Conditional Knockout Mouse
- Geraets, R. D. (2019). Neuronal Ceroid Lipfuscinosis: A Tailored Animal Model of CLN2 Disease and Evaluation of Select Personalized Therapies (Doctoral dissertation, ProQuest Dissertations Publishing).
- Zhao, M., Tao, F., Venkatraman, A., Li, Z., Smith, S. E., Unruh, J., ... & Marshall, H. (2019). N-Cadherin-Expressing Bone and Marrow Stromal Progenitor Cells Maintain Reserve Hematopoietic Stem Cells. Cell reports, 26(3), 652-669.
-
Li, C., Zheng, Z., Ha, P., Chen, X., Jiang, W., Sun, S., ... & Chen, E. C. (2018). Neurexin Superfamily Cell Membrane Receptor Contactin‐Associated Protein Like‐4 (Cntnap4) is Involved in Neural EGFL Like 1 (Nell‐1)‐responsive Osteogenesis. Journal of Bone and Mineral Research https://doi.org/10.1002/jbmr.3524.
-
Geraets, R. D., Langin, L. M., Cain, J. T., Parker, C. M., Beraldi, R., Kovacs, A. D., ... & Pearce, D. A. (2017). A tailored mouse model of CLN2 disease: A nonsense mutant for testing personalized therapies. PloS one, 12(5), e0176526.
-
Miller, J. N., Kovács, A. D., & Pearce, D. A. (2015). The novel Cln1R151Xmouse model of infantile neuronal ceroid lipofuscinosis (INCL) for testing nonsense suppression therapy. Human Molecular Genetics, 24(1), 185–196. http://doi.org/10.1093/hmg/ddu428.
For more journal references, please visit our comprehensive list of citations and reference publications.




