Newsletter
WHITE PAPER - Inquire for a free PDF! or DOWNLOAD
TARGATT™ Technology For Antibody Discovery and Screening - eBook Feature 2021
Applied StemCell's (ASC) TARGATT™ technology allows site-specific, irreversible gene insertion (up to 20 kb). ASC has taken its proprietary knockin technology and integrated the system into CHO and HEK293 cells. The TARGATT™ CHO and HEK293 Master Cell Lines have displayed high integration efficiencies and medium to high levels of protein expression. These cell lines have the potential to be used for bioproduction, to establish a mammalian cell-based library for antibody screening, and to construct a naïve IgG library system.
Read more about Applied StemCell's TARGATT™ technology and TARGATT™ Master Cell Lines in Informa Connect's eBook, Antibody Discovery, Selection & Screening.
TARGATT™ for Targeted Gene Insertion in Mouse Models
Applied StemCell Inc.’s proprietary TARGATT™ knock-in mouse technology enables highly efficient site-specific gene integration in mammalian cells and animals. This technology uses PhiC31 integrase to insert any gene of interest into a specific docking site that was pre-engineered into an intergenic and transcriptionally active genomic locus for guaranteed gene expression. TARGATT™ Technology can be utilized for generating mouse models for a variety of applications including reporter gene expression, gene knockdown, disease models and even site-specific Cre expression for advanced conditional knock-out models. We have compiled and reported data from our mouse model projects in a technical white paper titled TARGATT™ FOR TARGETED GENE INSERTION IN MOUSE MODELS. This paper highlights the advantages of this novel gene editing platform in generating site-specific knock-in mouse models, and Applied StemCell's expertise in successfully generating transgenic mice.
POSTERS - Inquire for a FREE PDF copy
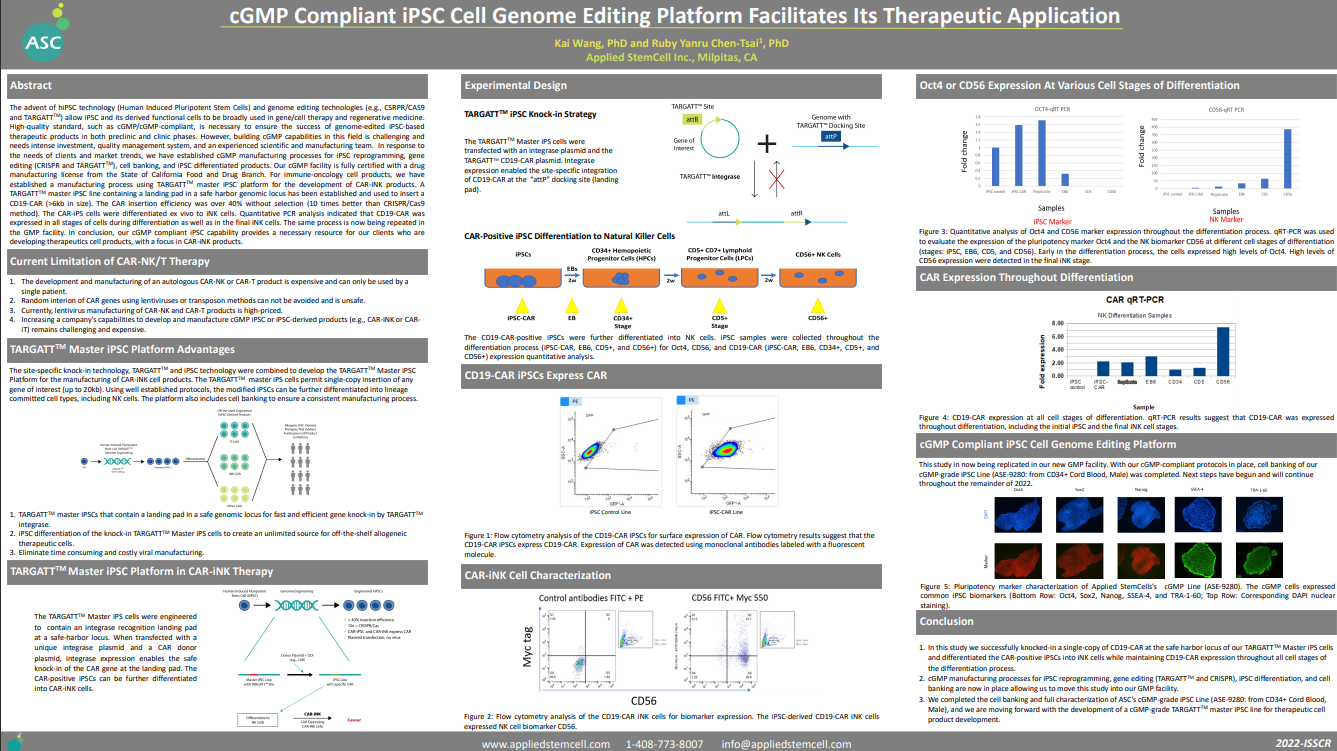
cGMP Compliant iPSC Cell Genome Editing Platform Facilitates Its Therapeutic Application >>>
Kai Wang, PhD and Ruby Yanru Chen-Tsai, PhD
Applied StemCell Inc., Milpitas, CA
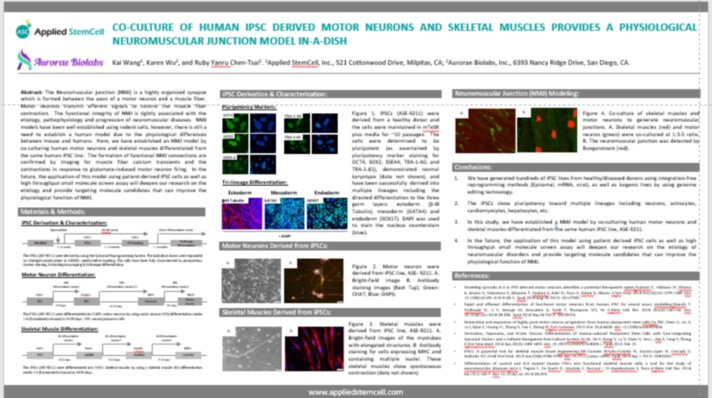
ISSCR 2020 - Co-culture of Human iPSC Derived Motor Neurons and Skeletal Muscles Provides a Physiological Neuromuscular Junction Model in-a-dish
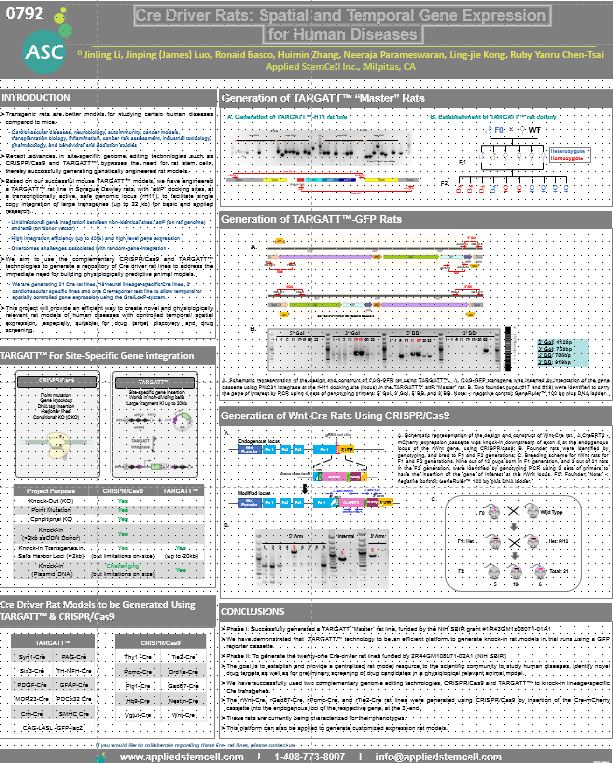
MBSJ 2018 - Cre Driver Rats: Spatial and Temporal Gene Expression for Human Diseases
The laboratory rat (Rattus norvegicus) is a central experimental animal in several fields of biomedical research, such as cardiovascular diseases, neurobiology, autoimmunity, cancer models, transplantation biology, inflammation, cancer risk assessment, industrial toxicology, pharmacology, and behavioral and addiction studies. A recent development of a TARGATT rat line provides a precise and convenient tool to generate KI rat models to facilitate researches in, but not limited to, the above-mentioned areas. The TARGATT™ site-specific gene integration technology uses bacteriophage phiC31 integrase to carry out efficient, unidirectional recombination between two non-identical sites, attP and attB.
The TARGATT™ rat system was engineered in Sprague Dawley rats with three attP docking sites at a transcriptionally active, safe genomic locus named rH11. It specifies following features: large cargo size, up to 20kb; single copy gene integration; high expression and high efficiency.
The TARGATT rats have been used to produce reporter model and Cre drivers, which have controlled temporal/ spatial gene expression. In parallel to a TARGATT™ mouse system, the rat system is expected to create a novel and much needed resource for the bio-medical community who employs rat models for their studies of human diseases.
WPC 2017 (#1) - Combined CRISPR - TARGATT™ Gene Editing Generates High Efficiency Knock-out, Point Mutation and Large Fragment Knock-in in Human iPS Cells
Gene editing in patient-specific induced Pluripotent Stem Cells (iPSCs) offers one of the most promising approaches for personalized therapy in regenerative medicine. CRISPR/Cas9 works most efficiently for gene knockout through the non-homologous end joining (NHEJ) pathway, and for point mutation/ correction through homology directed repair (HDR). However, efficiency for large fragment DNA knock-in through nuclease-mediated HDR is very low. Here, we present a few examples of knock-out, point mutation and large fragment knock-in with CRISPR/Cas9 in human iPS cells. We also present an alternative route to efficiently generate large fragment knock-in using a “Docking-site-ready” iPSC master line. The master cell line was generated by inserting an integrase recognition “attP” site into a specific genomic safe harbor locus using CRISPR/Cas9. We show data that with the TARGATT™ iPSC master cell line and integrase system, we can knock-in any transgene (up to 20 kb) at the preselected docking site with high efficiency (up to 30-40%), and without drug selection or cell sorting. These data support that CRISPR/Cas9 and TARGATT™ together provide a valuable new platform for efficient genome editing in human iPSCs.
WPC 2017 (#2) - Leveraging iPSC Technology for Disease Modeling and Drug Screening in Neurological Disorders
Human induced pluripotent stem cell (iPSC) technology offers the benefits of a cell line, coupled with the advantages of using human primary cells. We have developed a panel of iPSC lines for neurotoxicity assays and disease modeling applications. These cell lines include: 1) control lines, 2) patient-specific lines, 3) lineage-specific knock-in reporters, 4) isogenic controls of single and double knock-outs. We have also established scalable protocols for generating differentiated cells in an assay-ready format. Here, we describe the utility of these lines for neurotoxicity assays, including assays to determine the specificity of different neural cell types for a small range of chemicals and drugs from the Tox21 library, as well as for neuroprotective assays with dopaminergic neurons.
WPC 2017 (#3) - Using Continuous Bioluminescent Production to Perform Real-Time Metabolic Activity/Cytotoxicity Screening
Traditional bioluminescent imaging approaches based on firefly (luc) or Renilla (Rluc) luciferase provide single endpoint readings that can obscure the identification of compound-induced metabolic impacts, if they occur outside of the observational window. Gaussia luciferase (Gluc), which is secreted from cultured cells into the culture medium, allows assaying at multiple time points, but requires significant hands-on time that scales with each additional measurement. Here, we evaluate the use of a synthetic luciferase cassette (lux) that does not require a chemical substrate to produce its bioluminescent signal, and confers an autobioluminescent phenotype that can be tracked continuously using automated imaging systems. The synthetic luciferase was found to be compatible with a variety of cell types, including kidney (HEK293), breast (T47D and MCF-7), liver (HepG2), colon (HCT116), and pancreatic (AsPC-1) cell lines. The tested cell lines also demonstrated self-modulation of their autobioluminescent output signals in response to real-time metabolic dynamics, and all results correlated strongly with alternative assay systems. Upon treatment with the ATP synthase inhibitor, Oligomycin B, a rapid reduction in autobioluminescence was observed within 24 hours at concentrations ≥ 5 nM. Similarly, 10 µM methotrexate induced a ~30% decrease in signal in T47D and HepG2 cells and a ~50% decrease in HEK293 cells by 24 hours. HEK293 and T47D cells displayed half inhibitory concentrations (IC50) of ~47 nM and 71 nM following digoxin treatment, respectively, while HepG2 cells did not produce a sigmoidal dose-response. Similar responses were observed following doxorubicin treatment, which induced a sigmoidal dose-response by HEK293 cells (IC50 = 20 nM), but not by T47D or HepG2 cells. These results suggest that continuous luminescence can be used in high-throughput metabolic activity/cytotoxicity screening without the significant hands-on time or reagent costs associated with chemically-stimulated luciferases, while maintaining the unique metabolic backgrounds and differentially activated toxicity pathways of different cellular hosts.
ISSCR 2017 - CRISPR/CAS9 Platform Facilitates Efficient and Precise Disease Modeling in Human Induced Pluripotent Cell Line
The advent of hiPSC (Human Induced Pluripotent Stem Cells) and CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/ Associated Nuclease Protein-9) technologies has been heralded as a genetic revolution for gene therapy and regenerative medicine. Researchers are exploring the genetics and mechanisms of diseases by introducing mutations or correcting existing mutations in patient-derived iPSCs by using CRISPR/Cas9 techniques for a new era in personalized medicine. While CRISPR/Cas9 techniques has been widely and rapidly adapted for gene manipulation in various organisms in just a few years since its inception, the genome editing in hiPSCs has been slower to develop given the sensitive nature of these cells and the unique skills required to handle them. Furthermore, accurate disease modeling requires more dependency on homology directed repair (HDR) of the CRISPR/Cas9 nuclease system as well as precise single or bi-allelic manipulations. Various parameters affect the outcome of CRISPR-editing in iPSCs such as gRNA design, transfection of CRISPR elements, cell clone selection, etc. Here, we present some case studies to demonstrate how optimization of both iPSC and CRISPR/Cas9 protocol allows for efficient mutation models, including defined heterozygous and homozygous mutations, large fragment knock-in and conditional/ inducible expression models.
AACR 2017 - Using CRISPR/Cas9 to generate isogenic cell lines and reference standards for applications in cancer diagnostics
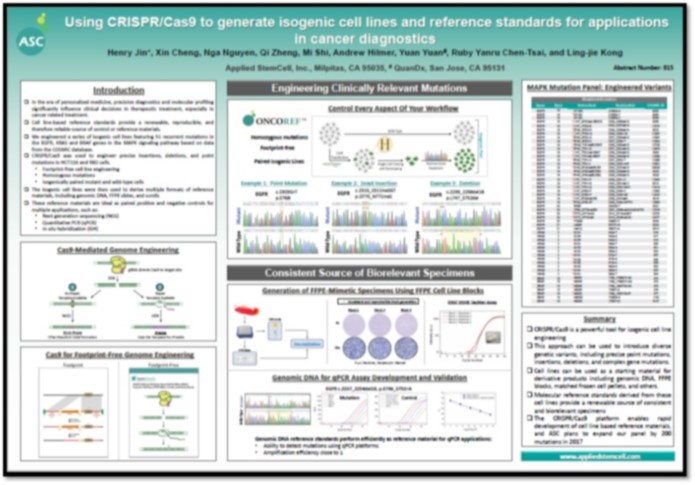
We are entering an era of precision medicine, in which there are significant initiatives to employ companion diagnostics and molecular profiling to inform clinical decisions in therapeutic treatment. In order for these initiatives to work as effectively as possible, there is a need for molecular reference materials that can help facilitate precision assay development. Cell-line based reference standards are ideal for such applications because they represent a reproducible and renewable source of control materials that can be employed for assay development, quality control, or lead compound discovery. With this in mind, we have used the CRISPR/Cas9 platform to engineer series of isogenic cell lines that feature 51 recurrent mutations in the MAPK signaling pathway. These cell lines contain footprint-free, homozygous mutations in the kinase domains of the EGFR, KRAS, and BRAF genes in HCT116 and RKO cancer cell lines, with 32 mutations in EGFR, 12 mutations in KRAS, and 7 mutations in BRAF. The mutations include precise deletions, point mutations and small fragment insertions, and these engineered cell lines have been further employed for the development of derivative reference materials, including genomic DNA (gDNA) and FFPE cell line blocks. Such materials can be employed as positive and negative controls in diverse assay platforms, including next generation sequencing (NGS), quantitative PCR (qPCR), in situ hybridization (ISH), and others. We present quality control data for these reference materials, which demonstrate excellent consistency and uniformity, as well as examples of the application of these materials in molecular assay validation. We conclude that using CRISPR/Cas9 technology to generate molecular reference standards is a very efficient and precise method in generating footprint-free, biorelevant mutations for companion diagnostic assays.
Discovery on Target 2016 - Genome Editing in Human iPSCs with CRISPR and TARGATT™ to Generate Cellular Disease Models
Site-specific Gene editing in patient-specific induced Pluripotent Stem Cells (iPSCs) offers one of the most promising approaches for the personalized investigation of the pathophysiology of disease and has great therapeutic potential in regenerative medicine. CRISPR/Cas9 works most efficiently for gene knockout through the non-homologous end joining (NHEJ) pathway, and for point mutation/ correction through homology directed repair (HDR). However, the efficiency for gene knock-in is not as satisfactory. Here, we present a few examples of gene knock-in, knock-out, and point mutation with CRISPR/Cas9 in human iPS cells. We also present an alternative route to efficiently generate large fragment knock-in using a “Docking-site-ready” iPSC master line. The master cell line was generated by inserting an integrase recognition “attP” site into a specific genomic safe harbor locus using CRISPR/Cas9. We show data that with the TARGATT™ iPSC master cell line and integrase system, we can knock-in any transgene (up to 20 kb) at the preselected docking site with high efficiency (up to 30-40%), and without drug selection or cell sorting. The TARGATT™-iPSC cell lines is a very efficient tool for reporter gene knock-in, generating inducible gene expression models and these data support that CRISPR/Cas9 and TARGATT™ together provides a valuable new platform for efficient genome editing in human iPSCs.
Symposium 2016 – Efficient CRISPR editing in blood lineage and blood-derived cell lines
This poster discusses details about ASC's milestones in editing blood-derived cell lines. The CRISPR/Cas9 technology has revolutionized genome editing in eukaryotic cells and embryos. Here we present our data based on our hands-on experience in blood-derived cell lines: (a) gRNA activity discrepancy in three major blood derived immune cell lines, Jurkat, K562 and TF1 cells; (b) Efficiency evaluation of the cloning-free CRISPR system by testing synthesized gRNAs with Cas9 expression plasmids; (c) Solutions for blood cells that are sensitive to transfection with DNA plasmids, such as the bone marrow derived BCWM.1 cells and the natural killer leukemia cells, KHYG1.
ISSCR 2016 – Gene editing and neuronal differentiation in human iPSC

Genome editing/correction in patient-specific induced pluripotent stem cells (iPSCs) offers one of the most promising approaches for personalized therapy in regenerative medicine. CRISPR/Cas9 has been widely used as an effective gene editing tool in the past two years for site-specific gene modification including insertion, deletion and nucleotide replacement. CRISPR works most efficiently in gene knockout through the non-homologous end joining (NHEJ) pathway. However for DNA knockin, the efficiency of CRISPR mediated homology-directed repair (HDR) remains low, limiting its application in therapeutics.
Our TARGATTTM technology allows site-specific gene insertion at a higher efficiency in a genomic safe harbor locus. We will discuss the results on knockin, knockout and point mutation efficiency in iPSCs using CRISPR/Cas9. We will also report using a “master” iPSC line for efficient site-specific insertion of large DNA fragments. With TARGATTTM and CRISPR, we are empowered with a complete set of genome editing tools to manipulate human iPSCs.
World Preclincal Congress 2016 – CRISPR-TARGATT™ gene editing in iPSC
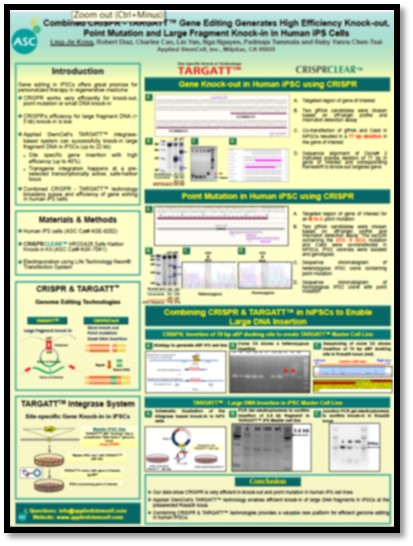
Gene editing in patient-specific induced Pluripotent Stem Cells (iPSCs) offers one of the most promising approaches for personalized therapy in regenerative medicine. CRISPR/Cas9 works most efficiently for gene knockout through the non-homologous end joining (NHEJ) pathway, and for point mutation/ correction through homology directed repair (HDR). However, efficiency for large fragment DNA knock-in through nuclease mediated HDR is very low. Here, we present a few examples of knock-out, point mutation and large fragment knock-in with CRISPR/Cas9 in human iPS cells. We also present an alternative route to efficiently generate large fragment knock-in using a “Docking-site-ready” iPSC master line. The master cell line was generated by inserting an integrase recognition “attP” site into a specific genomic safe harbor locus using CRISPR/Cas9. We show data that with the TARGATT™ iPSC master cell line and integrase system, we can knock-in any transgene (up to 20 kb) at the preselected docking site with high efficiency (up to 30-40%), and without drug selection or cell sorting. These data support that CRISPR/Cas9 and TARGATT™ together provides a valuable new platform for efficient genome editing in human iPSCs.
AACR 2016 - CRISPR editing in mouse
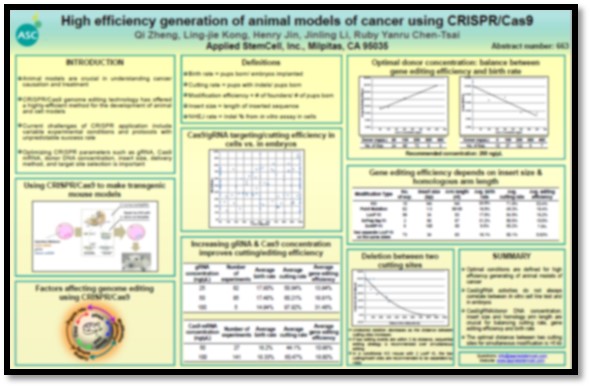
Applied StemCell is one of the first and most experienced provider of the CRISPR-Cas9 Technology for mouse model generation. Our expert team of scientists have extensively researched and upgraded the CRISPR system to optimize the efficiency of Cas9 cutting, modification efficiency and even improve birth rate of mice. The findings were presented at the prestigious AACR Annual Meeting 2016 in New Orleans in April. Some of key findings have been noted under technical details.
ISTT 2016 - TARGATT™ Rat Models

The laboratory rat (R. norvegicus) is a central experimental animal in several fields of biomedical research, such as cardiovascular diseases, aging, infectious diseases, autoimmunity, cancer models, transplantation biology, inflammation, cancer risk assessment, industrial toxicology, pharmacology, behavioral and addiction studies, and neurobiology. Until recently, the ability to create genetically modified rats has been limited compared to that in the mouse, mainly due to lack of genetic manipulation tools and technologies in the rat. The isolation and establishment of rat embryonic stem (rES) cells by Drs. Ying and Smith’s groups at the end of 2008 enhanced the capability of making genetically modified rat models. Recent advances in nucleases such as ZFNs, TALENs, and particularly, CRISPR have been successfully used to generate ‘knockout’ rat models by injecting gene targeting molecular complexes directly into an embryo. As a result, rat transgenics is becoming as precise and powerful as has been the case in mice, yielding better models of human diseases.
To facilitate the generation and use of rat models of human diseases, it is critical to develop systems that enable fast, efficient and precise introduction of exogenous genetic elements into the rat genome. We have previously developed an integrase-based TARGATTTM system for making site-specific transgenic mice. Here, we applied the same TARGATTTM approach to direct transgene integration at transcriptionally active locus of the rat genome. Integrases such as phiC31 carry out efficient, unidirectional recombination between two non-identical sites, attP and attB. We first identified a transcriptionally active locus in the rat genome and inserted attP sites at the locus using CRISPR. These attP-containing rats can be used as embryo donors for pronuclear injection of the transgene on an attB-containing plasmid. In the presence of integrase, recombination between attP and attB results in an insertion of the transgene precisely at the attP site in the rat genome. This technology allows fast, efficient generation of knock-in rat models containing any gene of interest with consistent, stable, guaranteed gene expression. Advantages of this integrase-based technology are: (1) Transgene integration happens at pre-selected and transcriptionally active locus; (2) Site-specific knock-in rat models are made by direct injection of the DNA into the rat zygotes, bypassing rES cells; (3) Gene integration efficiency is higher, but off-target events are lower compared to CRISPR. This TARGATTTM rat system offers a cost-effective method and valuable resources for the bio-medical community who employ rat models for their studies of human diseases.
Tricon 2016 – Gene editing in blood lineage cells
The bacterial CRISPR-Cas9 system has been tested successfully from test tubes to eukaryotic cells, and has emerged as a genome editing for multi-organisms. Here, we focus on examining the CRISPR system and compare its efficiency in blood-derived immune cell lines, Jurkat, K562, and TF1 cells. Eight gRNAs were tested among these 3 lines. We found the activities of the most gRNAs are similar in K562 and Jurkat cells, while seven out of eight gRNAs show significant lower activity in TF1 cells, suggesting that loci eligibility and efficiency of CRISPR are different not only from tissue to tissue but also from cells in the same lineage.
Guide RNA synthesis offers lower cost and efforts comparing to traditional plasmid-based sgRNA cloning approach. To evaluate this cloning free CRISPR system, we tested synthesized gRNAs with Cas9 expression plasmids. We found that when co-transfected with Cas9 DNA plasmids, synthesized gRNAs do not work, while co-transfection of U6-gRNA plasmids with Cas9 plasmid showed high efficiencies; Alternatively, transfection of these synthetic gRNAs in Cas9 expressing cells also led to effective Cas9 cleavage, suggesting that the synthetic gRNAs possess shorter lifespan and fail to synchronize with the Cas9 expression when using DNA plasmids.
ISTT 2015 - TARGATT™ Master Cell Line
TARGATT™ Posters
 TARGATT™-CellLines-CellLineEngineering-201806
TARGATT™-CellLines-CellLineEngineering-201806 CRISPR-TARGATT™-iPSCGeneEditing-WPC-201706
CRISPR-TARGATT™-iPSCGeneEditing-WPC-201706
 Production of site-specific transgenic mice using the TARGATT™ knock-in technology
Production of site-specific transgenic mice using the TARGATT™ knock-in technology